

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_086080 | OTHER | 0.997647 | 0.000119 | 0.002234 |
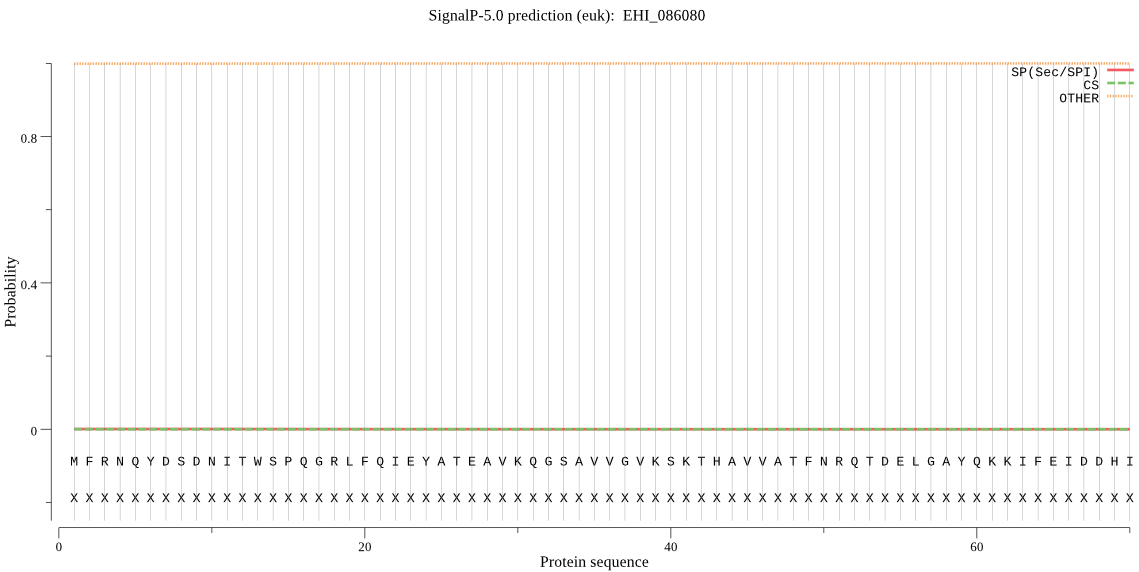
MFRNQYDSDNITWSPQGRLFQIEYATEAVKQGSAVVGVKSKTHAVVATFNRQTDELGAYQ KKIFEIDDHIGVAISGFCADARVITSQMRSECIDYSYVYGTPHPVEKLVKHVSDNAQVNT QKYGLRPYGVGLLVIGYDTKPRLFETSPSGLYYSYKAQAIGARSQSCRTYLEKHFESFKE LDEEQLIKQAILALRESLTTTNDKLSVLNCSVAIVGKDTPFRFITDKELQNQLDNLDTQQ MEMVQAEVD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_086080 | 177 S | KHFESFKEL | 0.995 | unsp |