

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
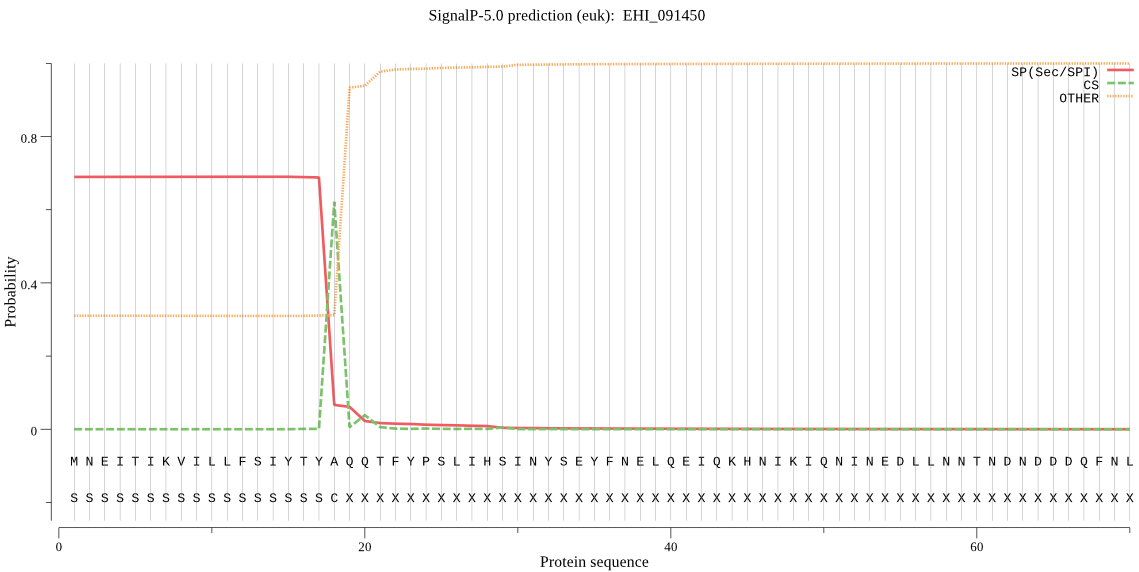
| EHI_091450 | SP | 0.083349 | 0.916527 | 0.000124 | CS pos: 18-19. TYA-QQ. Pr: 0.8618 |
MNEITIKVILLFSIYTYAQQTFYPSLIHSINYSEYFNELQEIQKHNIKIQNINEDLLNNT NDNDDDQFNLELPDEILWENFCKYFNKSYSTTAESNYRKNIFMEQIKIYRYFNSLSSSNI TAVFGITILSDKSLSEIGCSNPKSLKNSSIIFPPPILPQITDPPASFSRCGKYTLNNSNS MKTDDFCTDIYWSGCDGCYAFAAKEIAFISYKNQTLKSASINTNLYFDCSPEAKGCCGGD TEEILKDFPYVYFQNESSSLEPSQYLSCKHEKCFLQHPNGLVKNIIEFGSINKPEQFKSL LLVYGPFVSSIKLFNGIQGYKYGILDIEHCKNESVIASHSIVVVGYGTQNNSSYLIIKNW WNGWGEDNKGYMKISMTNSCGIGTQPEGKQPTNYIIQFYYCEGDSYCIDCDKETGKCLEC SYLSRLDKNGMCTRECPDDCLTCDKALEECFSCKNNAILQDGKCVIKCPKYCKTCERETL ECTSCQPSYILNGTECIYNCTDDCEICDPYNETCIKCKDKSELLNGYCNVLCPVGCKTCD RESQRCYSCKRFYRLNQYSCVSTCKDKCIDCDSSNTFCELCEVGYTLLNGECYSSVCKKN CRFCNITTPNCVECFLPYFSYNGDCQINSFLALIFLYIFITLSLLLNALL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_091450 | 548 S | QRCYSCKRF | 0.99 | unsp | EHI_091450 | 548 S | QRCYSCKRF | 0.99 | unsp | EHI_091450 | 548 S | QRCYSCKRF | 0.99 | unsp | EHI_091450 | 133 S | LSDKSLSEI | 0.991 | unsp | EHI_091450 | 543 S | CDRESQRCY | 0.996 | unsp |