

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
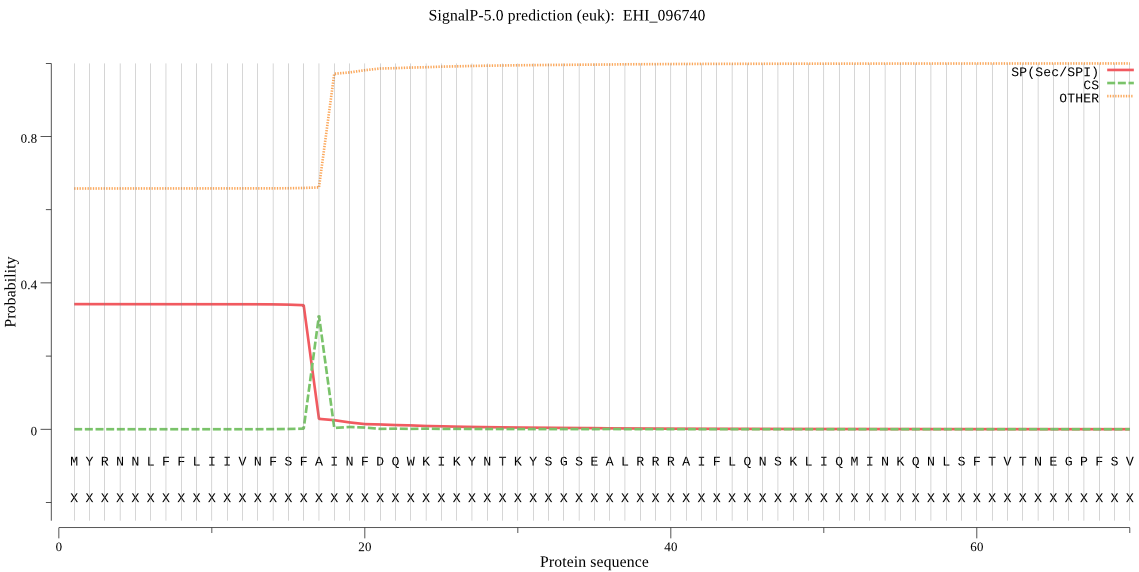
| EHI_096740 | SP | 0.062932 | 0.936057 | 0.001011 | CS pos: 17-18. SFA-IN. Pr: 0.7592 |
MYRNNLFFLIIVNFSFAINFDQWKIKYNTKYSGSEALRRRAIFLQNSKLIQMINKQNLSF TVTNEGPFSVLTNEEYRMLHHRIDIEKEIKQLKSHRMNLVKKMDNKEVLDSIDWRSEGKV TPVKNQRKCASCYAFGSIATIESLIMQETSIKEIDLSEQQIVDCSQGEYSNWGCTCGNVG NSFNYVRDHGILLERDYPYTGKANNCSIDGKKPVIKIKDYSFVFPQTEENLKIAVYHQPV AVSIDSSQLSFQFYEGGIYDEPNCKWVDHIVTVVGYGTTEEHQDFWVVKNSYGNEWG
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_096740 | 150 S | MQETSIKEI | 0.996 | unsp |