

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_097900 | SP | 0.000701 | 0.999298 | 0.000000 | CS pos: 15-16. ALA-EF. Pr: 0.9718 |
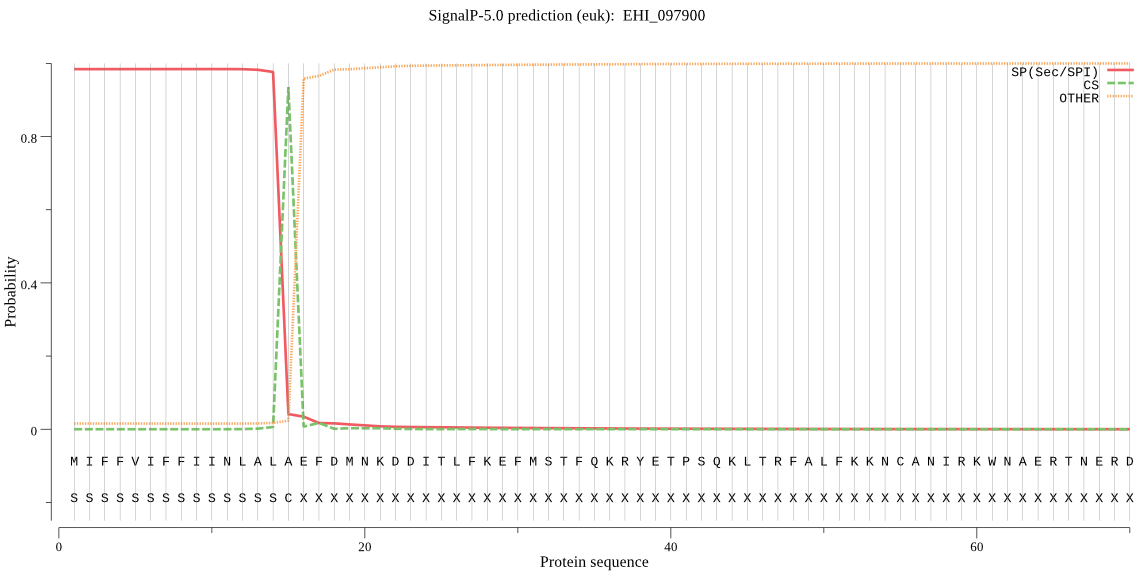
MIFFVIFFIINLALAEFDMNKDDITLFKEFMSTFQKRYETPSQKLTRFALFKKNCANIRK WNAERTNERDAHFGITSRTDKLPMEYGLSRNLNDLASKTKENDVIIDGGAYPPILPESDD LPESLSYCGDYVVNNTDHPKVNLCLTPYDQGSCGSCYAASTANLGQYLYANLSYYYNVGN QSNIVIKNFTAQRWIDQKNNFYVRRCCGGNTKMMLLTQPTFSTEYEYPYVDIHTSENDIN GCRNRGDQNPNVAIHLRTQKITVFGMDNTYSHSQKVTIIKKILHHYGPISVSILVDVNQT NNAIKMANYQGGIFKFPSTCNINKMGIDHQVIIVGYGVEDGEEYLIMRNSWGKWGEYDDG YMKISTETPLCGIGEIVENYSPSNYIIYAGNCILDRNCASCNSKTLVCSECKEGTTMDSR GMCLDNSYPSIPEDAVAPEEPNPDPDRLEDPPAEDSVNSLVIFSIISLIVLLI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_097900 | 42 S | YETPSQKLT | 0.994 | unsp |