

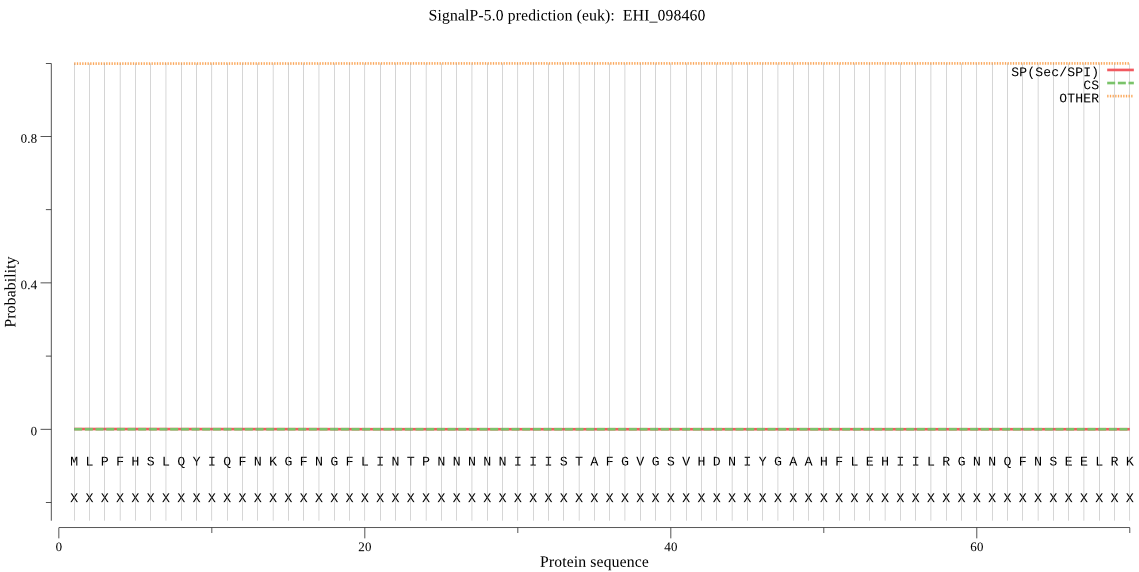
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_098460 | OTHER | 0.897743 | 0.003418 | 0.098839 |
MLPFHSLQYIQFNKGFNGFLINTPNNNNNIIISTAFGVGSVHDNIYGAAHFLEHIILRGN NQFNSEELRKMNNKQLLNISAITSREITKFQCITSPLYWKRDITTMMSLLFQPTFNNKQI IKENNIINSERQYVKMDKTQLLFQKKHELLFGNNTPFGHEIIGSEYSQKQIQFLELQTMY NKYYNSKNCCIGIICPYQYQTQLIKFINKNINQFYLKQYSKNQCFKSTHISQYKKPKIIS KDNYCIVTKGSFRNRLITDLINYKVSLNQESYHIPYHYYNLIISSSPLNLKFLQINSSLK ENYLNKVNEVFNTNPILLLDYYLMFELNFYYIHLN