

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_104610 | OTHER | 0.999963 | 0.000029 | 0.000008 |
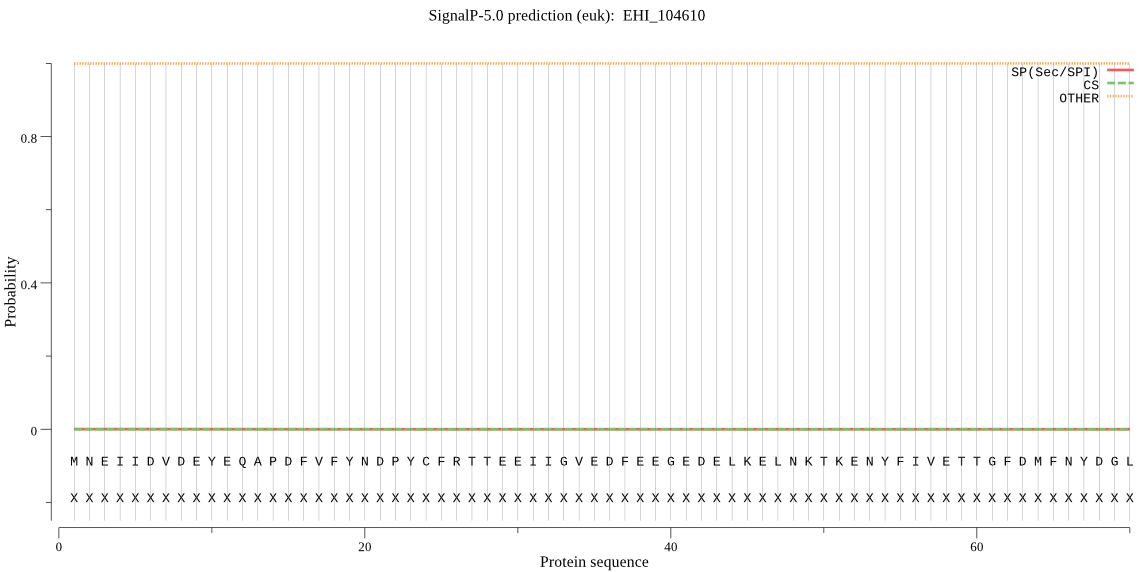
MNEIIDVDEYEQAPDFVFYNDPYCFRTTEEIIGVEDFEEGEDELKELNKTKENYFIVETT GFDMFNYDGLQLTQDITYFKIHFKEAVDYIKSIIQLDIIDIHDVFQNINKMIFRGHITNT IIEWSKRMTVCAGICYGKSNGCVIRLSEGILKYRSIYDVVTTIVHESIHAFLFRTKTRDN DSHGIRFHQWMTIINEKTPLHVTVYHTFYEECKNLHKHIWRCTGPCRNKPPFYGYVKRSM NRAPGPTDRWYEQHKKHCDGHFVKIDGPEFHEETLAKKPKRKERKSHTNKKEVKSKDLKD LFNELKKKKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_104610 | 155 S | LKYRSIYDV | 0.991 | unsp | EHI_104610 | 286 S | KERKSHTNK | 0.998 | unsp |