

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_114340 | OTHER | 0.999133 | 0.000816 | 0.000051 |
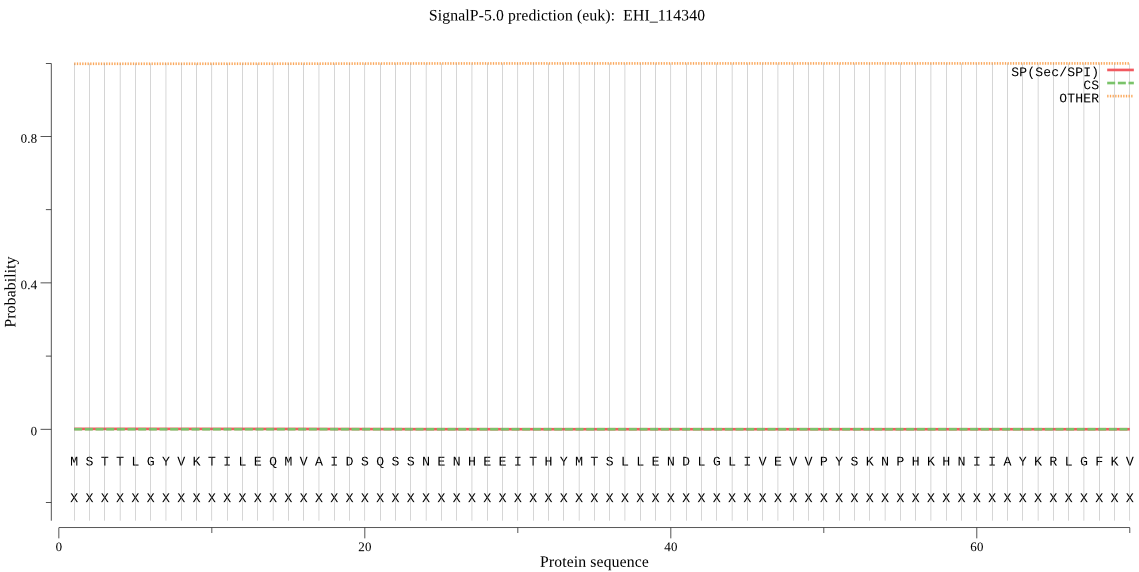
MSTTLGYVKTILEQMVAIDSQSSNENHEEITHYMTSLLENDLGLIVEVVPYSKNPHKHNI IAYKRLGFKVVLCGHLDTVNIGSGWTKEPLKCTTEVVDDKTYIYGRGTSDMKGGNAVIIA TLKRLIENNNNIDDIAIFFSTEEEVGVRGCQDFMVSHRHMFESVETFVVLEPTNLYVGSG QNGHYWVKYTCHGKSAHGCMPQDGLNAIDGMADLNCVLKEAITAPDLNGHVILNIGTIEG GDKVNIVPDLCIETVDYRFSPNVFDKHLEEIIKKVIDQMNEISYAQYEYEFISKISAIKC RSTNVLLNKLSEYMKSINQFKEVSLPYGSDAAVFKKEFPSSDFIIFGPGTVEQMHKTDEF VVLEHLNIAIEALVTALKGR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_114340 | 23 S | DSQSSNENH | 0.996 | unsp |