

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_117650 | SP | 0.011050 | 0.988943 | 0.000007 | CS pos: 15-16. SEG-SS. Pr: 0.6169 |
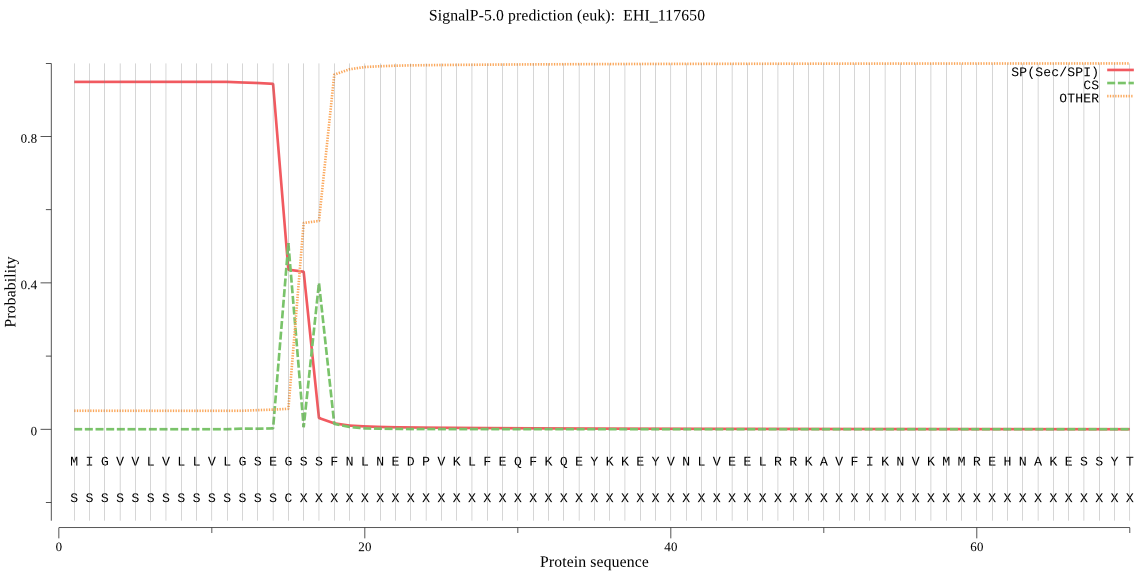
MIGVVLVLLVLGSEGSSFNLNEDPVKLFEQFKQEYKKEYVNLVEELRRKAVFIKNVKMMR EHNAKESSYTMGINKFADMEGNELLATSSKIQNNRRMGARRIQEKIKTEKVNLTKQTLKQ AVPANYTLCTSEAEYNYCGTNNIDQNVCGGCYAVGPAQHLSTIYAYLTKQADNNKPKRKM LSAQQLIDCNTESWGGCGGGFAENVLNSVDGIYYADDYPFIDGNTECEDPTDCTRFKHEC KKLNLGYPLKFATSNKFTHFANKNWEEIKEIIYTYHGFISAMKVTESLYRYSGGIYKSQS CTGQITDHVITVDGYGECDGHKFLWVRNSWGNDWGVEGGHFKIDWDTLCGINGIDHYEDG SQLQNNLYVEMELGDGDTADYNPQDGAGENKQDPIGCVVVSPTGDNDNKTTGIVITLFVL LMTFFI