-
-
-
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>EHI_121860
MYHTLERMYNISYFAFQWLGVAVFFLYLSSLILYVPPITSVSHNTTNMVKSFKKGSYSDF
TIDLDVDFTPSFNWNTKMIFVWVKASFTNKNVPYNTATVWDTMIRKKEKAHLHLTNERIE
YPLVSSYNSLLGKEVELTVEWMVVPWSGATTVEHGNSTKFVLPTEFSK
- Download Fasta
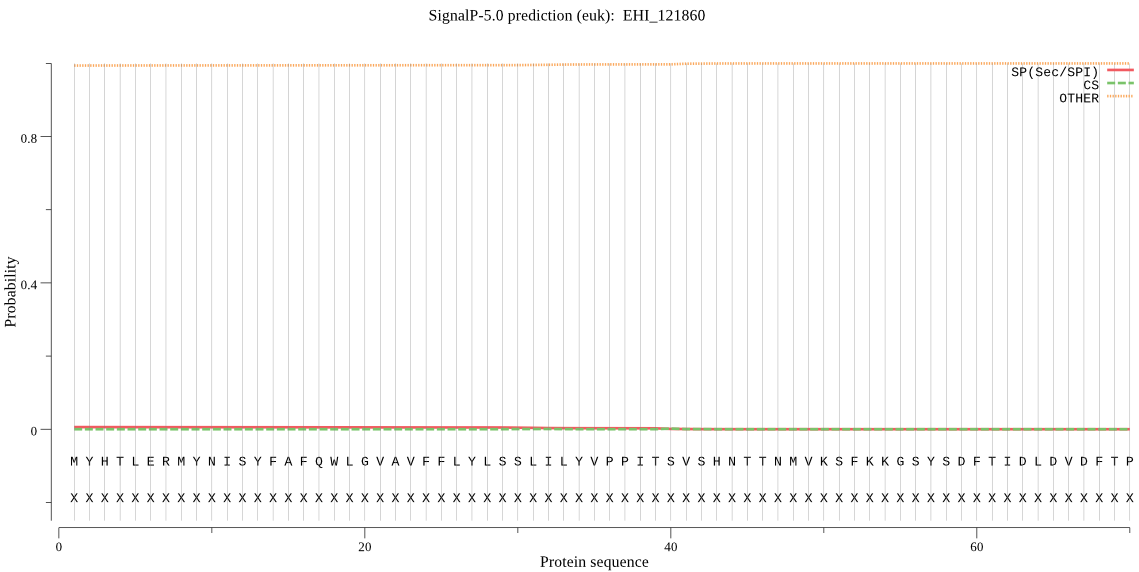
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/EHI_121860.fa
Sequence name : EHI_121860
Sequence length : 168
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.861
CoefTot : -1.510
ChDiff : 1
ZoneTo : 58
KR : 4
DE : 1
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.041 2.288 0.384 0.815
MesoH : 0.151 0.459 -0.292 0.322
MuHd_075 : 21.991 16.199 5.781 5.688
MuHd_095 : 29.072 22.772 9.161 6.539
MuHd_100 : 36.159 27.998 12.040 8.649
MuHd_105 : 40.309 28.409 13.020 8.983
Hmax_075 : 11.600 8.633 -1.458 4.680
Hmax_095 : 8.662 13.000 2.222 3.509
Hmax_100 : 17.600 16.500 3.053 5.870
Hmax_105 : 13.600 17.762 2.774 3.370
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.7581 0.2419
DFMC : 0.7464 0.2536
-
Fasta :-
>EHI_121860
ATGTATCATACTTTAGAACGTATGTATAATATTAGTTATTTTGCTTTTCAATGGCTAGGT
GTAGCAGTTTTCTTCTTATATTTATCATCATTAATACTATATGTTCCTCCTATAACTTCA
GTATCACATAATACAACGAATATGGTTAAGTCATTTAAAAAAGGTTCTTATTCTGACTTT
ACAATAGATTTAGATGTTGACTTTACACCATCATTCAATTGGAACACAAAAATGATTTTT
GTTTGGGTCAAAGCATCGTTTACAAATAAGAATGTTCCATATAACACAGCAACTGTTTGG
GATACTATGATCAGAAAGAAAGAAAAAGCACATTTACATTTAACAAATGAAAGAATTGAA
TATCCTTTAGTTAGTTCCTATAATAGTTTACTTGGAAAAGAAGTTGAGTTAACAGTTGAA
TGGATGGTTGTCCCATGGTCTGGTGCTACTACTGTTGAACATGGAAATTCTACTAAATTC
GTACTTCCTACTGAATTTTCTAAATAA
- Download Fasta
|
ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore |
|
EHI_121860 | T | 164 | 0.574 | 0.178 | EHI_121860 | T | 158 | 0.548 | 0.031 |
|
ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore |
|
EHI_121860 | T | 164 | 0.574 | 0.178 | EHI_121860 | T | 158 | 0.548 | 0.031 |
|
ID | Site | Peptide | Score | Method |
|
EHI_121860 | 56 S | FKKGSYSDF | 0.992 | unsp |


