

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_126170 | SP | 0.289441 | 0.695370 | 0.015189 | CS pos: 14-15. SFS-QK. Pr: 0.5513 |
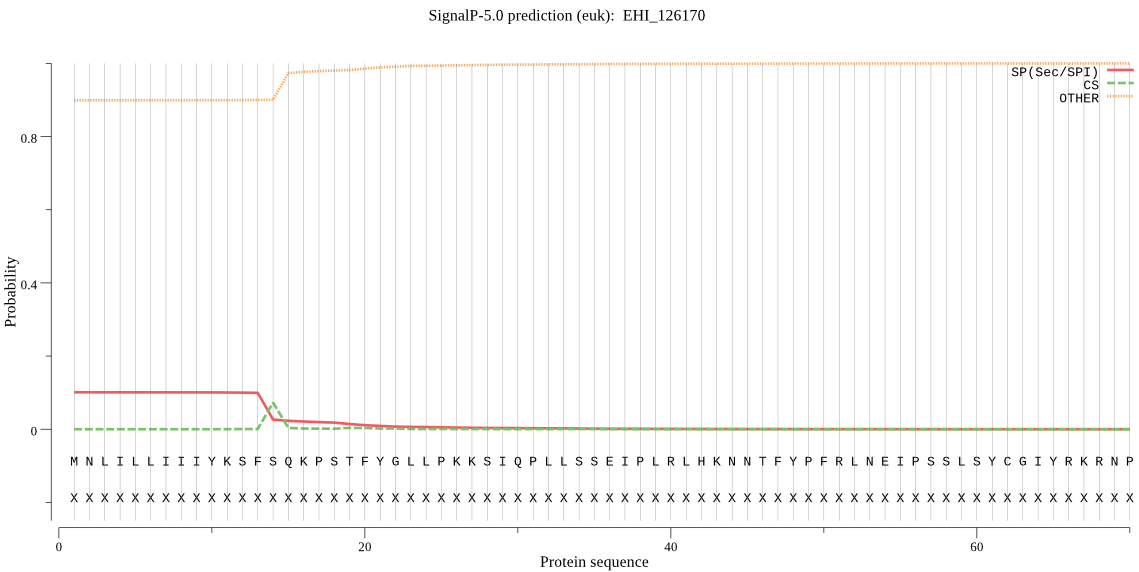
MNLILLIIIYKSFSQKPSTFYGLLPKKSIQPLLSSEIPLRLHKNNTFYPFRLNEIPSSLS YCGIYRKRNPHKTEDYCIKQTMNQGECGGCYAMALAQVLQAHYTHLTLKHQAFSTQQIID CSNNNGCSGGWPTSVLESLQYFVSEIEYPYRLITINGNSTNNYKKECIRGRRTKIHVSDY KEVIGPITFEQIKVLLIEHGPFIGMIYSNDQLRKYSGGILHLDCPVVPTLNHAIIVVGYG QENQEKYIIIRNSWGNSWGEMGYARISMNDLCGLDGIKYSDYPTILYLKTQCLFLFFSLN IITDNFIVPQQVYSNY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_126170 | 14 S | YKSFSQKPS | 0.996 | unsp | EHI_126170 | 267 S | YARISMNDL | 0.996 | unsp |