

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_130660 | OTHER | 0.997539 | 0.000153 | 0.002307 |
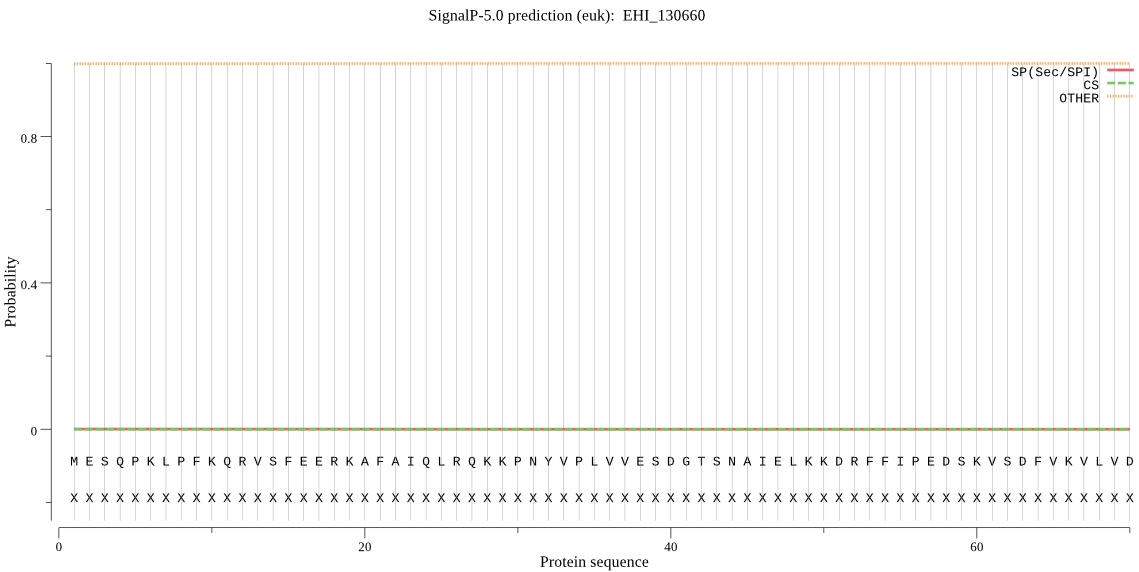
MESQPKLPFKQRVSFEERKAFAIQLRQKKPNYVPLVVESDGTSNAIELKKDRFFIPEDSK VSDFVKVLVDKYIETDGETPISTVSVKIQTPSKAIQPSNEDTIGSLYAMYQEEDGYLYFI VYRESVFGN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_130660 | T | 90 | 0.640 | 0.054 | EHI_130660 | T | 83 | 0.566 | 0.083 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_130660 | T | 90 | 0.640 | 0.054 | EHI_130660 | T | 83 | 0.566 | 0.083 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_130660 | 14 S | KQRVSFEER | 0.998 | unsp | EHI_130660 | 98 S | AIQPSNEDT | 0.996 | unsp |