

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
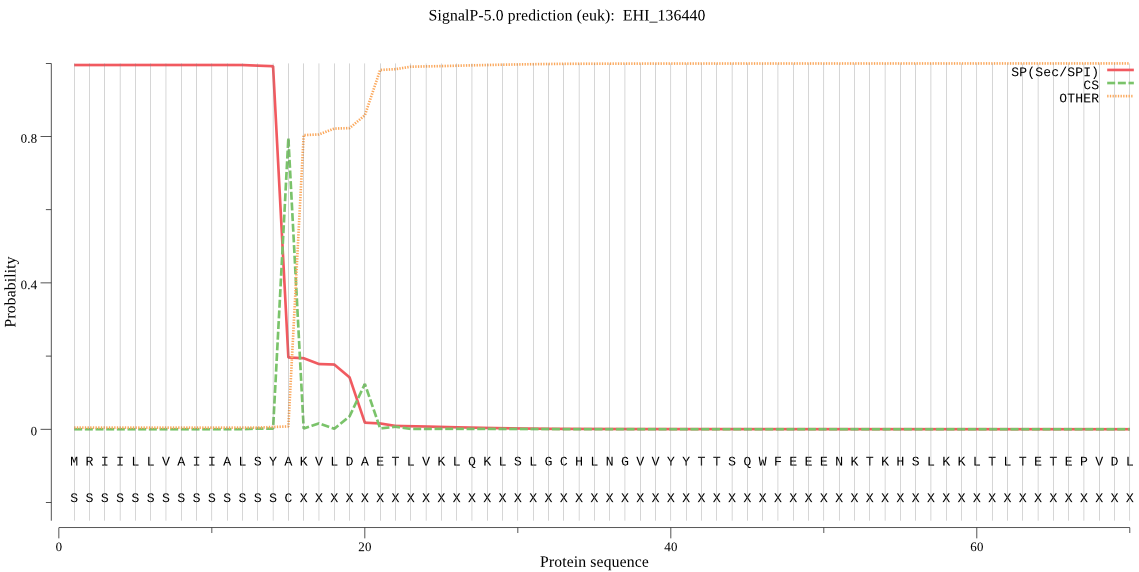
| EHI_136440 | SP | 0.000137 | 0.999856 | 0.000007 | CS pos: 15-16. SYA-KV. Pr: 0.8433 |
MRIILLVAIIALSYAKVLDAETLVKLQKLSLGCHLNGVVYYTTSQWFEEENKTKHSLKKL TLTETEPVDLIPYSEESFSNFVCLNEGVYLLHNGQIGKIENDKIVDITSTPISIDTFKMI SVGGKLRGLASMTVFPGMTLEESANKFIELEKQNYRVYDQLMIRRWDTYWDGQFQHLFTF KEEDDNSITFKDIMNDKKYDCPARPFGGNEEYDISPDGTMIAFSILLENPASSLDNNVYY ATFENPLEWNCITTNNKGYDNQPLFNNDGSLLYYLSMSAPKDESDKSVLKSYDFNQKVIR DITGNIDLSFSAPMKVVGNYMHLSTQIEGNVYIVGLDTSKSELTREDVNIITQKGTAGSF VFTTTGIIYEYNSFTLPQELFLYENKVIKQITHINQEVLSTIKFGEYKEIHYTGANNDQI HAFITYPPNMKTNTKYPVILYTHGGPESPWTNNFHYRWNPQVIAAQGYIVFAPNFHGSGS YGDAFLKAIRKNWGGWPFEDLMKGMDYLKTSEPLVDIDNACAMGASYGGYMMNWINSQNT GRFKCIICHDGIMDSEGSYYYMDEMYFLETEFGYPMYEDDTYYKKYSPLNYVNGYNTPQL TIHGGSDYRIDVVAGYQQFVTLQRKNIESKLVIYPEENHWVLRPYNSIDWHAQVFDWLAK YLPKK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_136440 | 480 S | HGSGSYGDA | 0.994 | unsp |