

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_138530 | OTHER | 0.999987 | 0.000007 | 0.000005 |
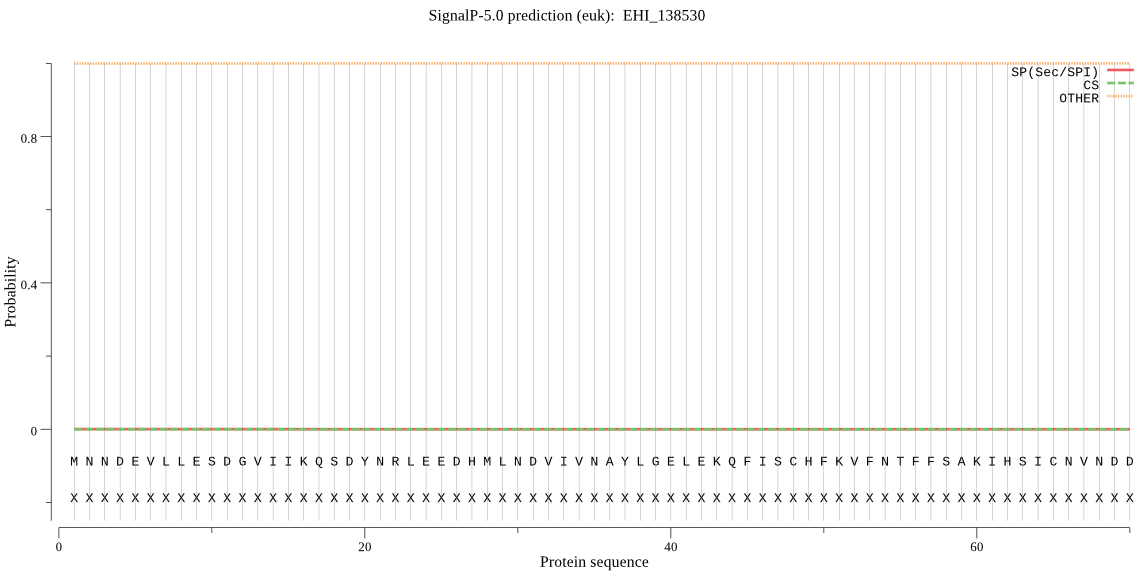
MNNDEVLLESDGVIIKQSDYNRLEEDHMLNDVIVNAYLGELEKQFISCHFKVFNTFFSAK IHSICNVNDDDLREQRYNQLVNWLKDDENLTDLRFLLFPCHYESHWFTVIVCNKTQKSQP QEDWERINSDEEGAVFEIEIGKEKSYCENYKKSDIGTELQDPLSQTLLDSPFKKQTTLSL NSEINTSPNLFFSSQNSTELKKNLSQLRDASISIEPCCEDYIDSPCILVIDSLKSISQTN ELTNNILEFIRWEYKRKEKEKKWDEEWEQHKRILSLDVPQQNNGVDCGVFMLYFIRKFME YTPSDGTKFKQIVGDIDPKKERLYIKNVIRELINRKTNKVMLD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_138530 | 221 Y | CCEDYIDSP | 0.991 | unsp | EHI_138530 | 221 Y | CCEDYIDSP | 0.991 | unsp | EHI_138530 | 221 Y | CCEDYIDSP | 0.991 | unsp | EHI_138530 | 129 S | ERINSDEEG | 0.993 | unsp | EHI_138530 | 145 S | GKEKSYCEN | 0.992 | unsp |