

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
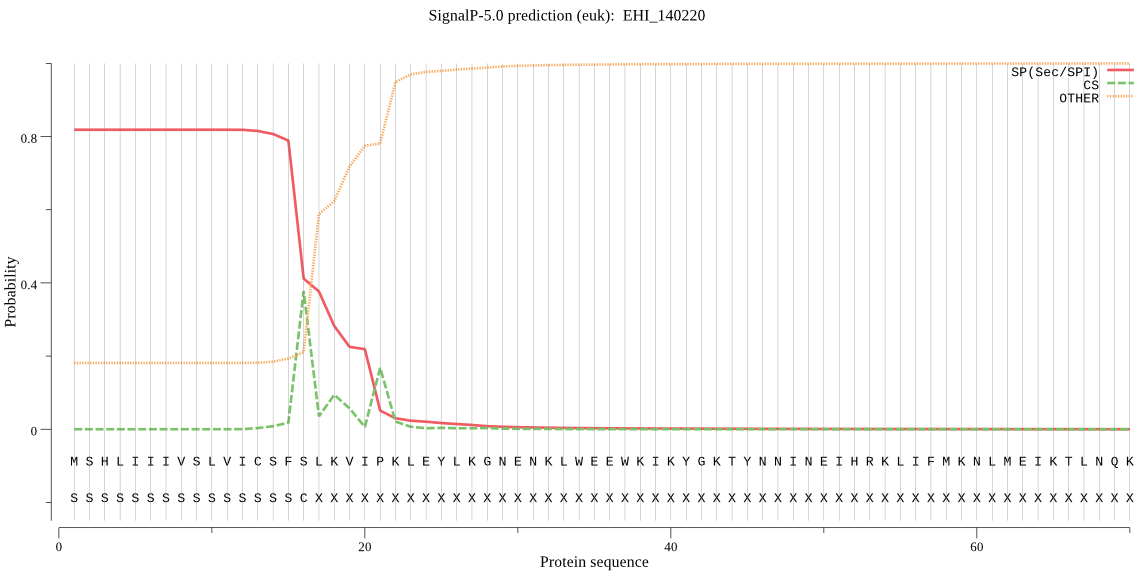
| EHI_140220 | SP | 0.026960 | 0.972902 | 0.000138 | CS pos: 21-22. VIP-KL. Pr: 0.4052 |
MSHLIIIVSLVICSFSLKVIPKLEYLKGNENKLWEEWKIKYGKTYNNINEIHRKLIFMKN LMEIKTLNQKREKDVDAYFDLNQWSDLSNQEFEDLMLMKKPKRSNAKELHDTINITIPYP KGPVPINYSACNQKTLFGKLNPGEIDFCNGIEFDQQSCGSCYCVSNALALQLKWANLTYL RDGKPQYKMFSPQQLLDCEVGGYRCAGGYADSVLDFSHYVSTIDDYPYYSGREPSKRMAC VKGKRTPIKLSYTIFDSAEDINIIKPIIHHYGGFVSCVYPKYWTAYRGGILRGLKCEKGV VTTHVVGIVGYGIEDGIEYVVVRNSWGKNWGLGGYIKLGADSLCGIGGNDGGDVPVSVVL HVDFSDVEYGPYGEFRNNTDVPQRLYNESAEANESSSNIESSNDSSNESQSLNSRQSEES LVVEDSSSVEPNIRPNNENHLRSITIYVLYVLVGISIITMVVAVIILHRSIIFN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_140220 | 235 S | GREPSKRMA | 0.992 | unsp | EHI_140220 | 235 S | GREPSKRMA | 0.992 | unsp | EHI_140220 | 235 S | GREPSKRMA | 0.992 | unsp | EHI_140220 | 397 S | NESSSNIES | 0.99 | unsp | EHI_140220 | 417 S | NSRQSEESL | 0.996 | unsp | EHI_140220 | 221 S | SHYVSTIDD | 0.994 | unsp | EHI_140220 | 230 S | YPYYSGREP | 0.993 | unsp |