

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
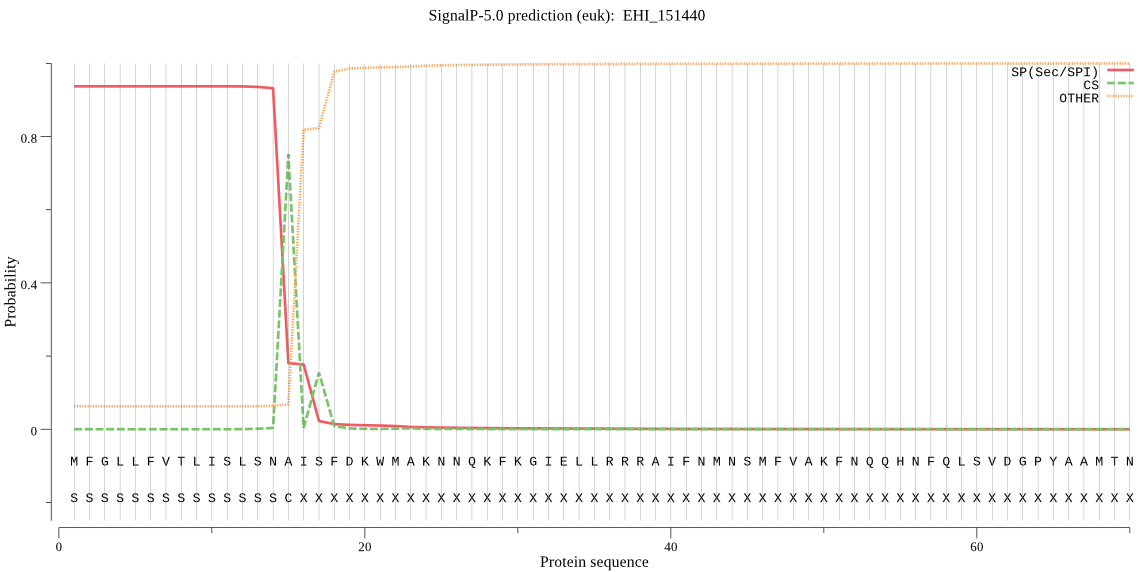
| EHI_151440 | SP | 0.001373 | 0.998523 | 0.000104 | CS pos: 15-16. SNA-IS. Pr: 0.7685 |
MFGLLFVTLISLSNAISFDKWMAKNNQKFKGIELLRRRAIFNMNSMFVAKFNQQHNFQLS VDGPYAAMTNAEYNTLLKARTVKNVNAPVRKAIKGDIPTAIDWRAEGKVTPIRDQAQCGS CYSFGSLAAIESRLLIGGSQTYNADNLDLSEQQIVDCSNKNNGCNGGSILYVFAYTKRNG VMQEKDYPYTATNGTCQYDADKIIVKNAGQVIVEQRNEVALVEAIAEGPVAVAIDAGQAS FQLYKSGVYDEPKCKKVILNHAVCAVGYGSQDGQDYYIVRNSWGTSWGMDGYILMSRNKN NQCGIANDAIYPTGVTEVKK