

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_152220 | SP | 0.002637 | 0.997346 | 0.000017 | CS pos: 22-23. KWA-YN. Pr: 0.6827 |
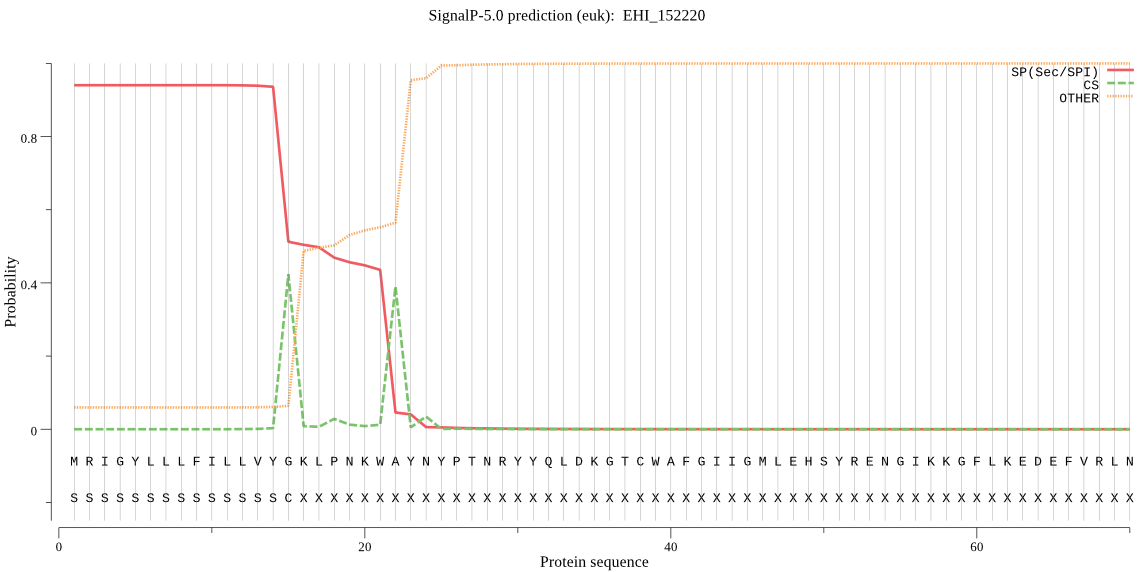
MRIGYLLLFILLVYGKLPNKWAYNYPTNRYYQLDKGTCWAFGIIGMLEHSYRENGIKKGF LKEDEFVRLNVQSFGILMVDACKKYPSVCNTPGDDVIFGSTEGGEINWFYSFPFLYDKIL PSAVCPYTATVDTQFECNGMDEALKTNPIKFNITEMLTTYNEEQTKELLLKVKIPIGFGA LIHDAKYYLPCTEEYKNFCDESVYNVIECPENMKYLAEKCAYIVMPMYSTDGEFNYHNEI EPEGGHAMVTIGYNDEYVTHEGCKGGFILKNSWNDTVYGPSIANTARGVRGSHSVKYFMN QLTAEEERKVCPNAQDPMNWYVCDDACVTNEELHKTIVNELYQAYKLQCVNPEEHFCETG YDYYLTELKADSKSPMNHYYIATFTKYDSTGKKVDTITLPSLPTSIIGMIFTPVEEQLIK LHDSEEFCGHYMFPYCILNKHLPFWGGYVGSHFEIEWDDRSYLINKDKYPEFDYKFIEES TFHQNLNLVDQKAGVPFLNERI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_152220 | 374 S | ADSKSPMNH | 0.991 | unsp | EHI_152220 | 374 S | ADSKSPMNH | 0.991 | unsp | EHI_152220 | 374 S | ADSKSPMNH | 0.991 | unsp | EHI_152220 | 50 S | MLEHSYREN | 0.993 | unsp | EHI_152220 | 272 S | ILKNSWNDT | 0.992 | unsp |