

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_153620 | OTHER | 0.994094 | 0.001743 | 0.004162 |
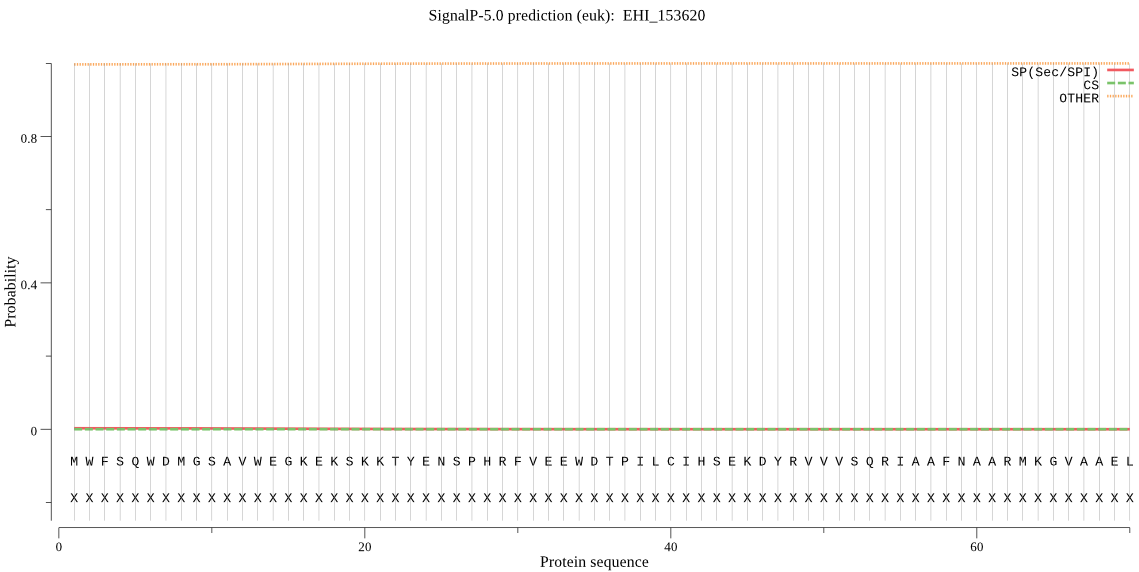
MWFSQWDMGSAVWEGKEKSKKTYENSPHRFVEEWDTPILCIHSEKDYRVVVSQRIAAFNA ARMKGVAAELLYFPDENHWVVKAQNGMLWQRTFFRWLRRWLE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| EHI_153620 | 43 S | LCIHSEKDY | 0.997 | unsp |