

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_159610 | SP | 0.002292 | 0.997677 | 0.000031 | CS pos: 13-14. ACA-NE. Pr: 0.6383 |
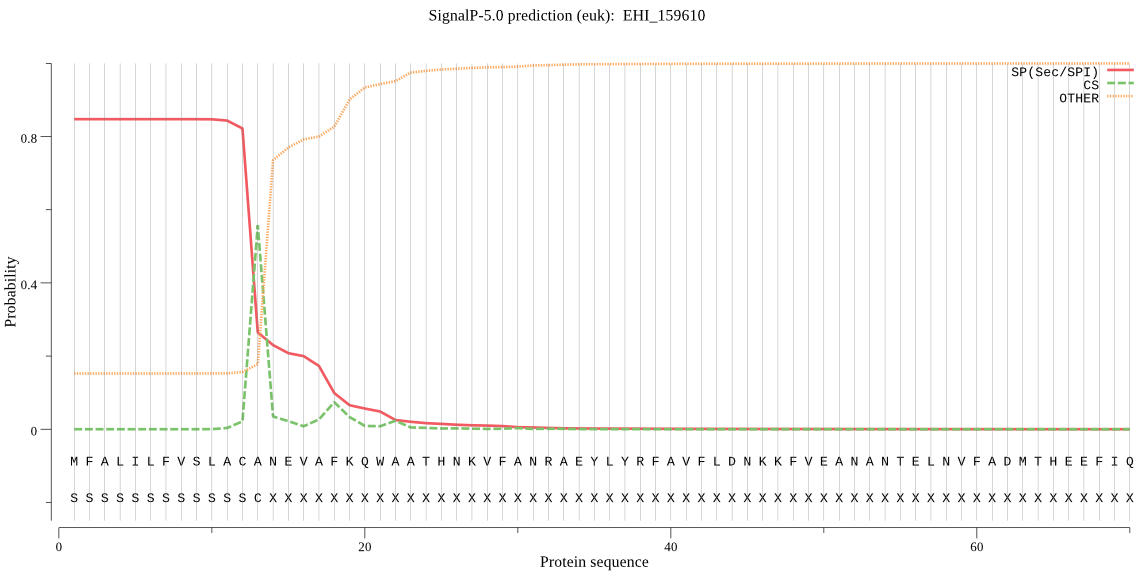
MFALILFVSLACANEVAFKQWAATHNKVFANRAEYLYRFAVFLDNKKFVEANANTELNVF ADMTHEEFIQTHLGMTYEVPETTSNVKAAVKAAPESVDWRSIMNPAKDQGQCGSCWTFCT TAVLEGRVNKDLGKLYSFSEQQLVDCDASDNGCEGGHPSNSLKFIQENNGLGLESDYPYK AVAGTCKKVKNVATVTGSRRVTDGSETGLQTIIAENGPVAVGMDASRPSFQLYKKGTIYS DTKCRSRMMNHCVTAVGYGSNSNGKYWIIRNSWGTSWGDAGYFLLARDSNNMCGIGRDSN YPTGVKLI