

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_168240 | SP | 0.019610 | 0.979839 | 0.000552 | CS pos: 13-14. AYA-TN. Pr: 0.5011 |
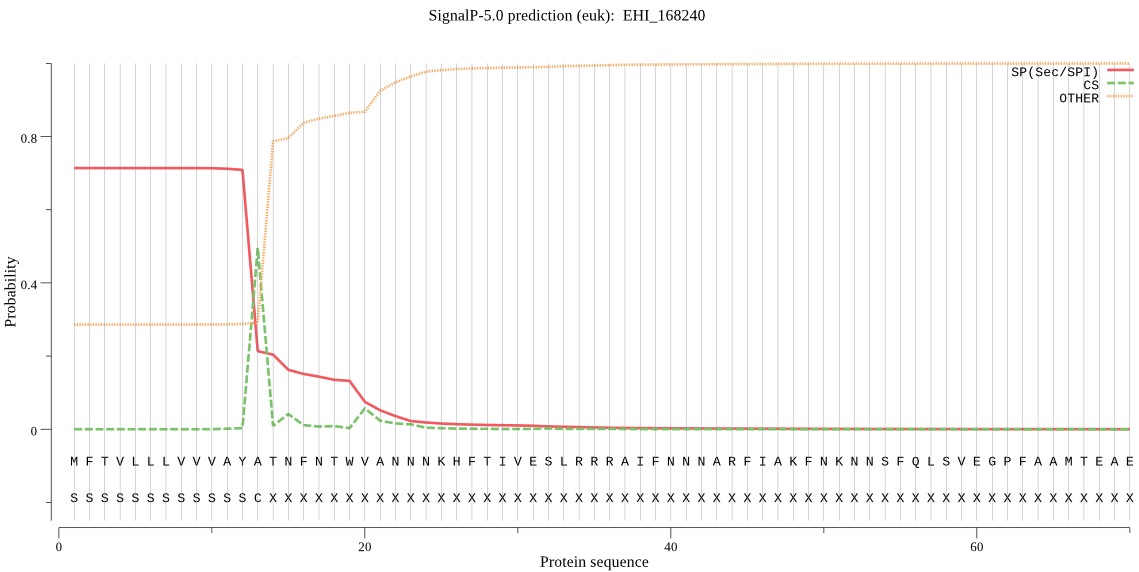
MFTVLLLVVVAYATNFNTWVANNNKHFTIVESLRRRAIFNNNARFIAKFNKNNSFQLSVE GPFAAMTEAEYNSMLKPFVIDKQHEEIVYDSRGDVPESVDWRAKGKVPAIRDQASCGSCY SFASVAAIEGRLLVAGSKKFTVDDLDLSEQQLVDCSVSVGNKGCNGGSLLLSFRYVKLNG IMQEKDYPYVAAEETCTYDKKKVAVKITGQKLVRPGSEKALMRAAAEGPVAAAIDASGVK FQLYKSGIYNSKECSSTQLNHGVAVVGYGTQNGTEYWIVRNSWGTIWGDQGYVLMSRNKN NQCGIASGAAYPVGVADA