

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
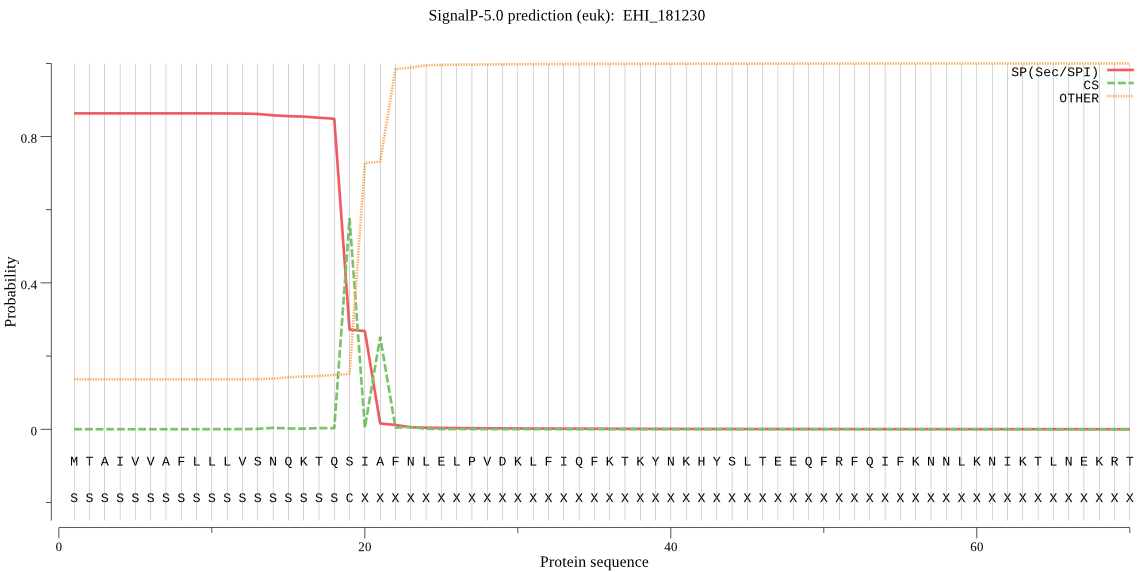
| EHI_181230 | SP | 0.129882 | 0.869808 | 0.000310 | CS pos: 19-20. TQS-IA. Pr: 0.6573 |
MTAIVVAFLLLVSNQKTQSIAFNLELPVDKLFIQFKTKYNKHYSLTEEQFRFQIFKNNLK NIKTLNEKRTQPSDAFHDINMYTDLIDEELPISKGMAIPVSSYDNEHFNSKELKKVEKPW NEVPPLPSGDNLPQNYAFCGEYVSKNTDRPKVDLCGEVFSQQNCGGCWAVSLANLAQYLY SNLTYQRYGCIKTPPKFSAQRFLDVTKTKATKRCCGGHPRDAVNAVKSFSLDADYPFVDG PNKNVCTIRGDQNPNVQIQMAITGYKTFGISKDQNMVLLLKKLLHHYGPFLVSVKVSGTG FSSYSSGIFSFPSSSICDTSSSSFMTDHEVVLMGWGIENGVEYFIIRNSWASGWGEGDNY IRIKTTNLCGIGWRIGDYVYPLNMIVYANTCKLDKYCSSCNISSNICSSCVSGYYLNSNN KCVKNTAKLVKYHSNSTYVQFYNHTI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_181230 | 101 S | AIPVSSYDN | 0.994 | unsp | EHI_181230 | 315 S | FPSSSICDT | 0.993 | unsp |