

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
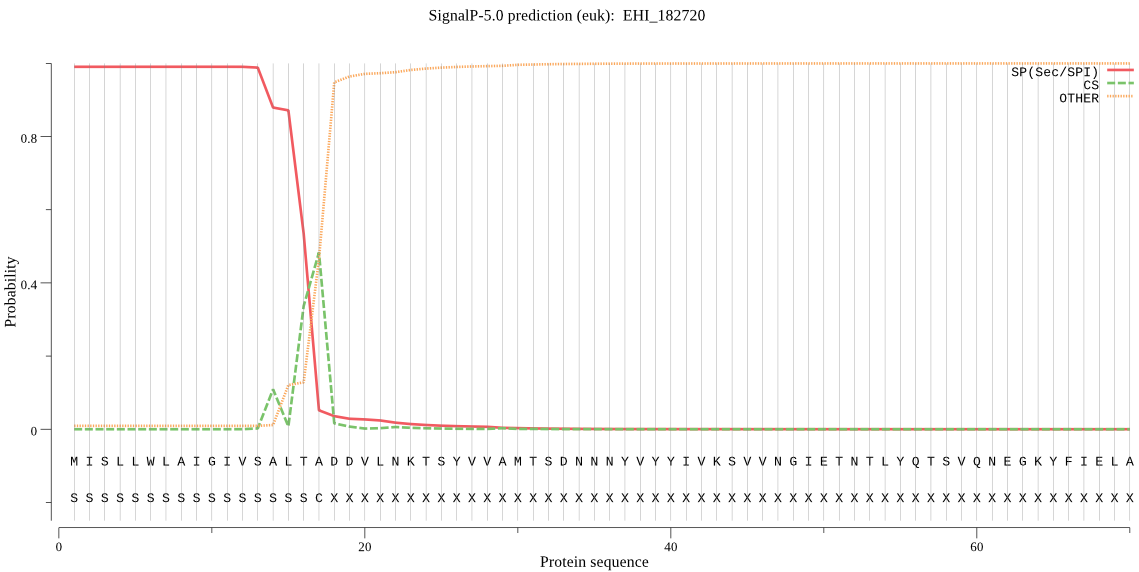
| EHI_182720 | SP | 0.001239 | 0.998749 | 0.000012 | CS pos: 17-18. LTA-DD. Pr: 0.4854 |
MISLLWLAIGIVSALTADDVLNKTSYVVAMTSDNNNYVYYIVKSVVNGIETNTLYQTSVQ NEGKYFIELAQGVGNELYYSVAHHSIFYTKCDGICSLYSLDSNMQEYQVTKDIAVSLPVI IKEHLYFVSPIFFGMTVQDSLDLQKKLANRKQTSHHVFTQTPFGEGSSYVTTNDDGTPQY KHLISSAITVDENGRIIVGQYTDVSNEVGNVISYDISDDESIVVFEAKSTASASDKKNYL KSSIYTISVGSDQATCISCNYPGNQIEPKATKTGKYIVFMRTTNTLEKNPAKYFVLYDLE DGSYRADVTLNEFTTEKYLIGKHSNEETITVFVINNRREALAVRKAVYSFESKAWNIEEV VVEGAVEMMAEVPCQDESSCLFIAQSFQYQPSEIYYCDEAMKLTKLTKLNDYTSYNLQKP ILVNPMGSLHDRIQSLLYLPSNNEKAPILLVVHEEEEGTVVDGFNKIMNPYVYTNEGYAV VMINSHVSKGFGERMRVKGSNDINVIIEDIQAVINELEKYPVDTSKIAVVGVFGASKIIT GLLDAKLPIVLAAVHAPILSIRSQYYTDAKYTLEYTFEGADYDNSYWYSHNDAMKNVASY NVPTLISYGDLDFVVDKSQPISLFQALQRRGIKSRLLRFPYSTNVLTKNEDIKVFYEQYI RWFNENMKN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_182720 | 232 S | KSTASASDK | 0.994 | unsp | EHI_182720 | 324 S | IGKHSNEET | 0.994 | unsp |