

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
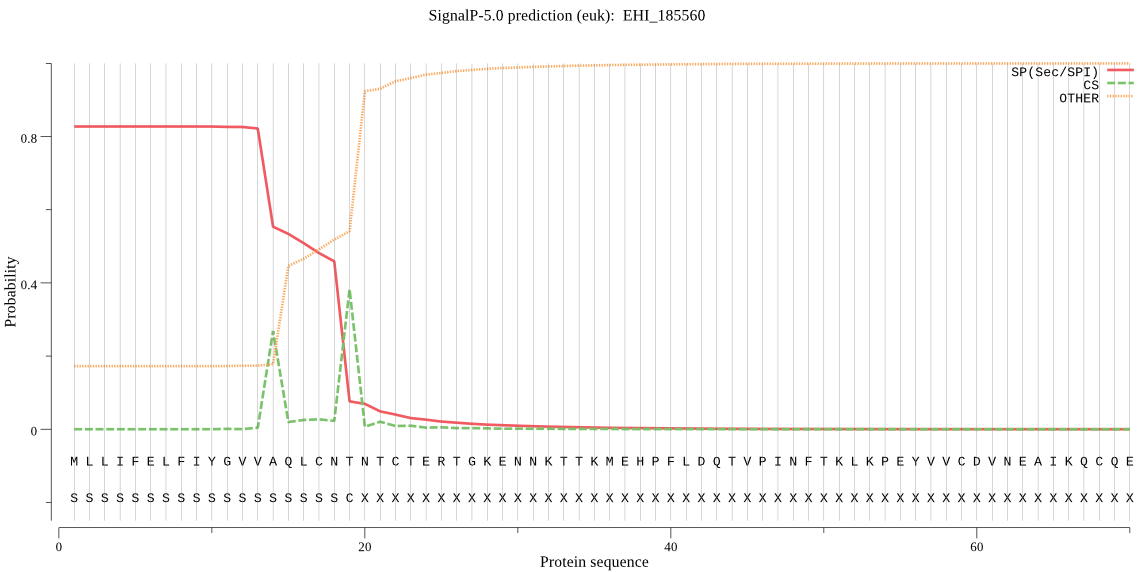
| EHI_185560 | SP | 0.122497 | 0.872936 | 0.004567 | CS pos: 19-20. CNT-NT. Pr: 0.6063 |
MLLIFELFIYGVVAQLCNTNTCTERTGKENNKTTKMEHPFLDQTVPINFTKLKPEYVVCD VNEAIKQCQEQINQIKNIKFEEVSFENVVRAFDNATIVIDNVMLKLEVLVSCRSDIEEYR KAYDEILPKVTEFRAALHSDEVMFSLVKKVYENKEKLEDDQKRLVEEKYEDFIEGGANLN KDDKTKLVELTKDITRLTKDFSNNLIDAVDKYTMYIEDKSKLEGIPENITEMYKKEAEER KKEGYIITLKVPSYFPAIKYCKNEELRKELFIQYNKLCSEGQFDNTKLIQEIIEKRQKYA ELLGFHDFSDLVTKRRMVKNGTNAIEFINQLHDKVQSFFYKEEDELKEFKKEKSGNNQLE PWDVNYYIELMKQEKYSFNEEEFREYFTEESVFGGLFEIYKRLYGVSFKLNKEVQGWHDD VKYYEVLEKDGSVIGGVYFDLYPRKGKRSGAWEEMLRVSYENENGTFIKPLGIVCTNITP PINGIARLNHDEVETIFHESGHLMHTLFSKQRYASLAGTNVAWDFVELPSQIMENWTFER EALDLFSQFGNVKPIPKELFDKMWKAKNYMSAYYTIRQLSFGKIDLEIHRNYLTSGLSLD DFIYQKEKDYKYHFNIKTFSIIRTFQHLFSNFIGYACGYYSYKWAESISADAFEYFKENG IFNQSIASKFRTNILSMGNSKPADELYRNFRGRNPDFTALLRRSGLVEKK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| EHI_185560 | 459 S | MLRVSYENE | 0.992 | unsp | EHI_185560 | 580 S | IRQLSFGKI | 0.991 | unsp |