

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| EHI_197490 | SP | 0.008003 | 0.991984 | 0.000013 | CS pos: 14-15. INA-IS. Pr: 0.7766 |
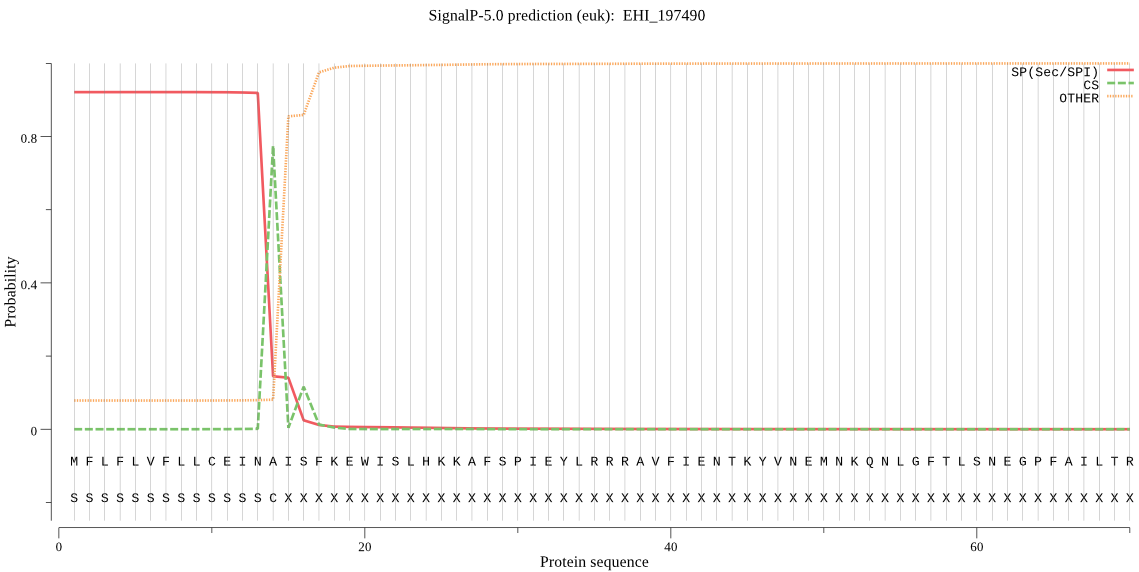
MFLFLVFLLCEINAISFKEWISLHKKAFSPIEYLRRRAVFIENTKYVNEMNKQNLGFTLS NEGPFAILTREESVAIAQGIHIDKSDLEQYKPSKREMVEAIDYRNIQGKSYMTPVKDQGN CGSCYAFSSVALMETAVLLSYDDLSPSNYALSTAEIVSCCYDPSECRGCEGGSIGGALKY AQDNGMQSESSFPYKPFEQHCLQNQKVMKVKKYTHSDTKGDDEKVRSEILSYGPVGSAMD ASRSSFLLYHGGIYNDKKCRSDKSTIAVVIVGYGIDKNNGKYFIVRNSWGPYWGEQGYFR ISSDNNLCGLSNDIYYIESIERLK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_197490 | 261 S | KKCRSDKST | 0.994 | unsp | EHI_197490 | 261 S | KKCRSDKST | 0.994 | unsp | EHI_197490 | 261 S | KKCRSDKST | 0.994 | unsp | EHI_197490 | 29 S | KKAFSPIEY | 0.994 | unsp | EHI_197490 | 145 S | YDDLSPSNY | 0.991 | unsp |