

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
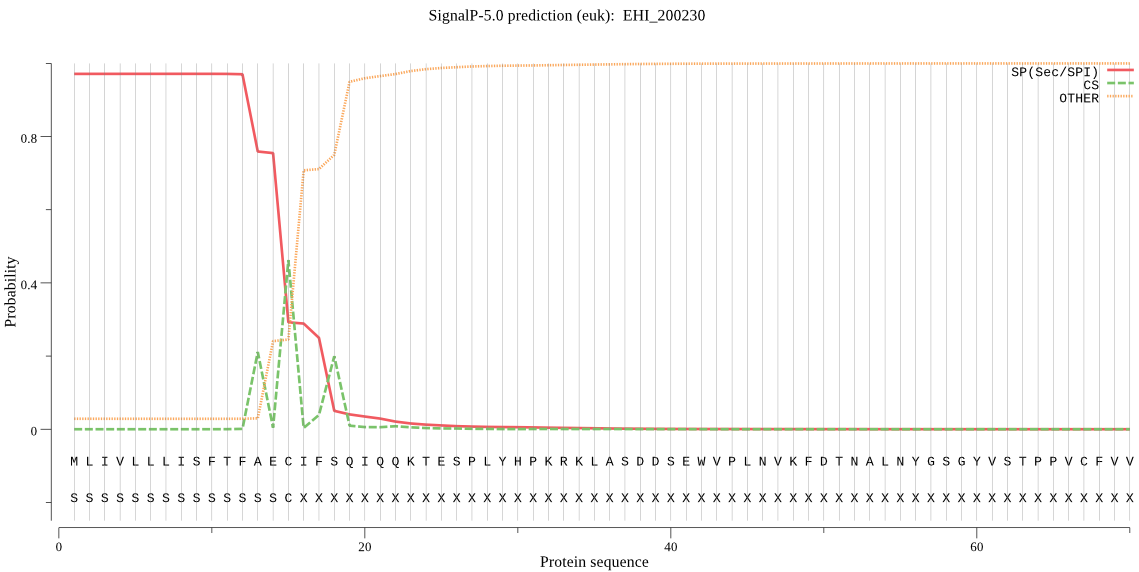
| EHI_200230 | SP | 0.003527 | 0.996458 | 0.000015 | CS pos: 15-16. AEC-IF. Pr: 0.4728 |
MLIVLLLISFTFAECIFSQIQQKTESPLYHPKRKLASDDSEWVPLNVKFDTNALNYGSGY VSTPPVCFVVSGTCTQDNLLTQEKKAYIIRIIDEVQRLIKKYFRVHKGTLATLKSSIKDK DRCGEISSIKDSSIADDIGMVMYVTAHPIESQTVLAYAASCGSAADASSTNPNNQKRNIF GYTNINPANLDVSEGKFRINAHTVLHETMHAMGFVSPTGMMSISKGRGTETVPVVTSEKV LKVAREHFGDNSISYVEFEDGGGSGTAGAHWEKRVLYNEIMTGTASSYSVISNFTLAYFE DLGTYSVNYSAAEPLTWGKGMKKDFFKCSNWPTQAPYYGETQARGCTPDRGAIGICDTSV RKDLPKIYQNYEDPTKGGMIELMDYCIHTTLVSGGQCYEKSVLSTENIASLSFLDRGSSY GKDSRCFSSSLMKYSIPISDFSCYRVKCVDRGYRVNVNGNWILCPSGDSISVTGYGGVIT CVNQSELCNGEVEEWPDIWRTDPVKGKAGSIVTLIGDYFSHMKKVYVGETEQTQFSIDNS NQVRVKIQFNDPFVNLIQLLSDGYVTVDIKIGDGNDINAVYQNFKLQVELVEVVQNVGQW LYKNLFFTVGIIIFLIFLVLLFGFIITKRIIYRRAKQVARNLV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHI_200230 | 404 S | KSVLSTENI | 0.991 | unsp | EHI_200230 | 404 S | KSVLSTENI | 0.991 | unsp | EHI_200230 | 404 S | KSVLSTENI | 0.991 | unsp | EHI_200230 | 116 S | TLKSSIKDK | 0.997 | unsp | EHI_200230 | 133 S | IKDSSIADD | 0.991 | unsp |