-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00000555
MHLRNGRVIGPTPRETSTMSVNKMPSGTPAHHRHATPKPTVAERPHGHKEAPPQVIPMTN
VMPGDSPFVTWDQFQSAASVIRTVIGGTVESGATIPILPEDGSNMQAFLLRIECRYTQVG
LEPRE
- Download Fasta
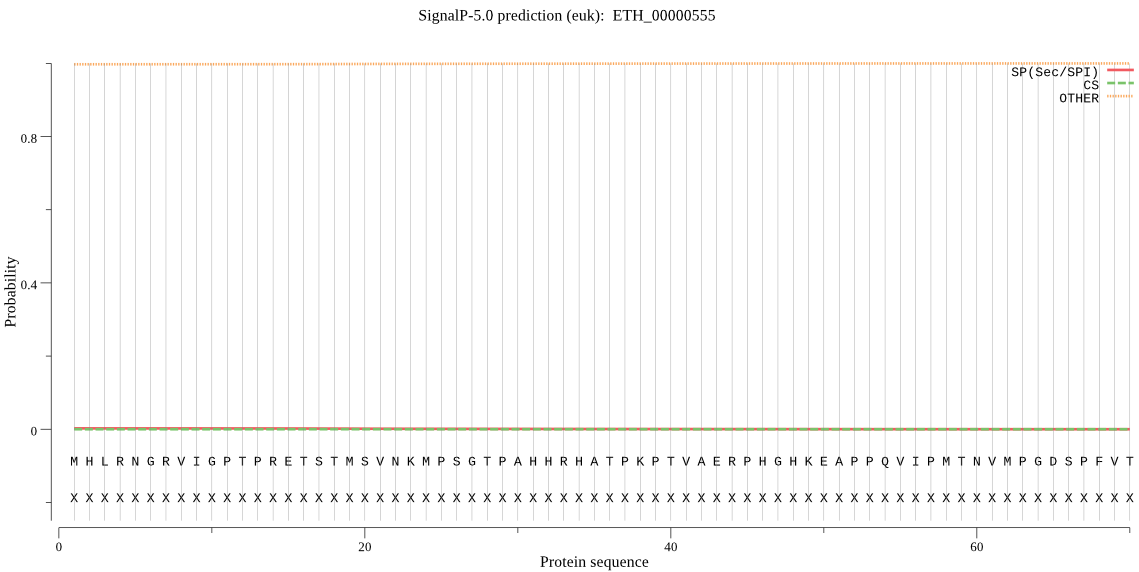
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00000555.fa
Sequence name : ETH_00000555
Sequence length : 125
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.963
CoefTot : -2.530
ChDiff : 1
ZoneTo : 42
KR : 6
DE : 1
CleavSite : 35
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.029 0.982 0.118 0.514
MesoH : -0.395 0.149 -0.342 0.202
MuHd_075 : 30.914 18.112 8.946 6.693
MuHd_095 : 38.716 19.762 9.932 7.260
MuHd_100 : 34.495 12.625 7.365 6.641
MuHd_105 : 39.398 15.865 8.217 8.453
Hmax_075 : 3.733 2.567 -1.755 2.543
Hmax_095 : 11.300 7.000 1.299 3.380
Hmax_100 : 8.700 2.800 1.225 3.220
Hmax_105 : 11.900 6.700 1.261 3.510
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.0743 0.9257
DFMC : 0.2133 0.7867
This protein is probably imported in mitochondria.
f(Ser) = 0.0714 f(Arg) = 0.0952 CMi = 0.35129
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>ETH_00000555
ATGCATCTCCGCAACGGACGCGTTATCGGCCCCACACCGCGGGAGACTTCTACAATGTCA
GTGAACAAAATGCCGAGCGGGACGCCGGCCCATCATCGTCATGCTACTCCGAAGCCCACC
GTGGCAGAGCGGCCGCATGGGCATAAGGAGGCGCCCCCGCAGGTGATACCTATGACCAAC
GTTATGCCAGGGGACAGTCCCTTCGTCACGTGGGACCAATTCCAGTCCGCTGCCAGTGTG
ATAAGAACTGTTATAGGGGGGACCGTGGAATCCGGCGCAACGATTCCCATACTACCAGAA
GATGGCAGCAATATGCAGGCCTTTCTGCTGCGCATCGAGTGCCGGTACACCCAGGTGGGA
TTGGAGCCGCGCGAGTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00000555 | 79 S | QSAASVIRT | 0.995 | unsp | ETH_00000555 | 79 S | QSAASVIRT | 0.995 | unsp | ETH_00000555 | 79 S | QSAASVIRT | 0.995 | unsp | ETH_00000555 | 12 T | VIGPTPRET | 0.991 | unsp | ETH_00000555 | 36 T | HRHATPKPT | 0.996 | unsp |


