-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00000920
MDAASSASFWTFSASMASSGAHTSYAFAGGERALEELRCGASSAAVWFCFLGDFDSGASR
VYGFPVRLAAAGSSSSSSNGSSSNGSSSNGSSSSKKRLRISLDLALAPREPLFLALQNVD
GTQAVPAAAAAPAAAAAAAAAAAAAPQPFLAIQNPTQAPPLLNGGALLLVHPDDIAVPPS
AAAAAAAAEAAAAAGEKPECANGATPLEEAAAAAAAAEAAQQKASLESREGQRPTKRPRA
FTPAAAPAATPAAAPAADGEAAAAAAKPEAVDAAGEVKAGAATAAAAPAAAAAGPESEAA
KSLDGKPPAATTAATAAAAAAKKVDLTLEEELQKERMRKRTAGIICFRWDKKGQRLLLSN
ARGIWACDAPMALQPLLQQTSLPPSNSAAAAAAEQDSPSSLSAQVLALRNLVQQQPAAAS
AAAAAAAGSPSSSSSGPREQPQPASPAPPPAAATAAAAAAAAAKEAWIHPSGRFGCSPAA
VCILERSPHEPPVLDCTCSSGTQQNR
- Download Fasta
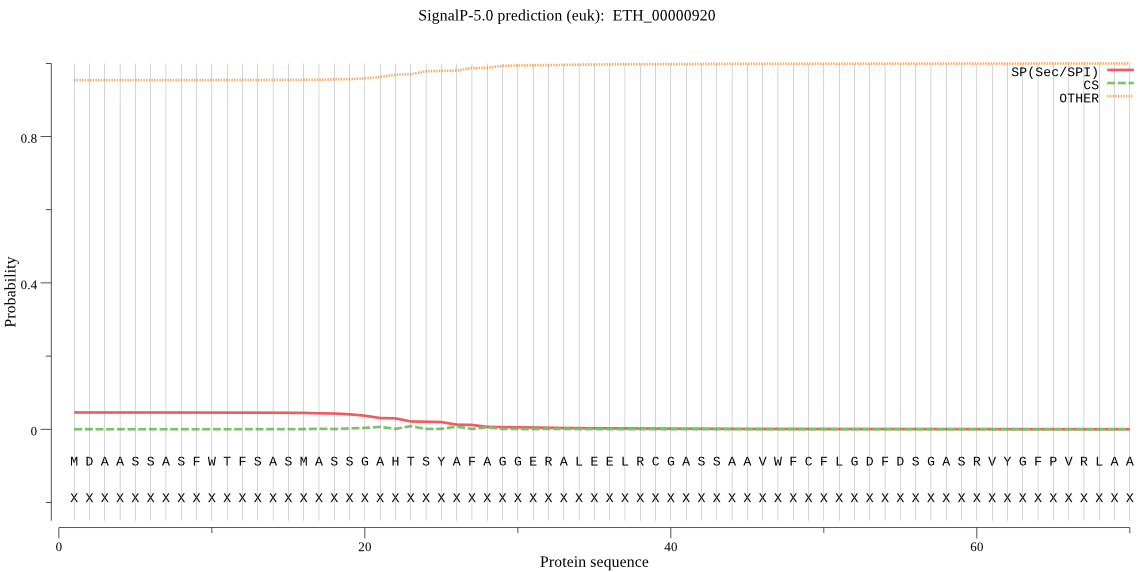
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00000920.fa
Sequence name : ETH_00000920
Sequence length : 506
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.739
CoefTot : 0.640
ChDiff : -3
ZoneTo : 30
KR : 0
DE : 1
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.535 1.600 0.225 0.591
MesoH : 0.567 0.828 -0.090 0.426
MuHd_075 : 8.328 9.300 2.328 2.936
MuHd_095 : 12.186 9.885 3.645 3.354
MuHd_100 : 10.331 8.639 3.104 2.857
MuHd_105 : 10.177 9.309 3.318 2.585
Hmax_075 : 10.600 9.700 0.871 4.500
Hmax_095 : 17.000 7.962 1.265 3.850
Hmax_100 : 14.200 4.800 1.215 3.390
Hmax_105 : 12.483 10.733 1.605 3.520
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9301 0.0699
DFMC : 0.8455 0.1545
-
Fasta :-
>ETH_00000920
ATGGATGCTGCTTCTTCCGCTTCCTTTTGGACATTTTCCGCCTCCATGGCGTCGTCGGGG
GCCCATACCTCATACGCATTTGCAGGGGGGGAGCGCGCACTGGAGGAGCTGCGCTGCGGG
GCCTCTTCAGCTGCTGTTTGGTTTTGCTTCCTGGGGGACTTCGACTCGGGGGCTTCTCGG
GTCTACGGCTTCCCTGTGCGGCTTGCTGCTGCAGGCAGCAGCAGCAGCAGCAGCAACGGC
AGCAGCAGCAACGGCAGCAGCAGCAACGGCAGCAGCAGCAGCAAGAAGAGGCTGAGGATA
TCGCTGGATTTGGCCCTTGCGCCTAGAGAGCCTCTCTTTCTAGCGCTGCAGAACGTAGAC
GGGACGCAGGCCGTGCCTGCAGCAGCAGCAGCACCAGCTGCAGCAGCAGCAGCAGCAGCT
GCTGCTGCTGCTGCGCCGCAGCCTTTCCTGGCCATACAAAACCCCACACAGGCGCCACCG
CTGCTGAACGGCGGCGCTTTGCTGCTCGTACACCCCGACGACATCGCAGTGCCCCCGTCA
GCAGCAGCAGCAGCAGCAGCAGCAGAAGCAGCAGCAGCAGCAGGAGAGAAGCCCGAATGT
GCAAATGGGGCCACGCCGCTGGAAGAGGCCGCTGCTGCTGCTGCTGCTGCAGAAGCAGCG
CAGCAGAAGGCCTCTTTAGAAAGTCGAGAGGGCCAAAGGCCCACAAAGAGGCCCAGAGCC
TTCACACCTGCTGCTGCTCCTGCTGCCACTCCTGCTGCTGCTCCTGCTGCTGACGGCGAG
GCAGCAGCAGCAGCAGCAAAACCGGAAGCAGTAGATGCAGCAGGAGAGGTTAAAGCGGGC
GCAGCAACAGCAGCAGCAGCACCAGCTGCTGCTGCTGCTGGGCCAGAAAGTGAGGCGGCG
AAGTCGTTGGATGGAAAGCCACCAGCAGCAACAACAGCAGCAACAGCAGCAGCAGCAGCA
GCAAAGAAGGTGGACTTGACCCTGGAGGAAGAGCTTCAGAAGGAACGCATGCGCAAAAGA
ACAGCAGGCATTATTTGCTTCCGATGGGACAAAAAGGGGCAGCGGCTGCTGCTCTCCAAT
GCTAGGGGTATTTGGGCATGCGATGCTCCTATGGCGCTGCAGCCGCTGCTGCAGCAAACC
TCGTTGCCCCCTTCAAACTCAGCAGCAGCAGCAGCAGCAGAGCAGGACAGCCCCTCCAGC
CTCTCTGCACAGGTTCTGGCACTACGGAACCTTGTGCAGCAGCAGCCTGCTGCTGCGTCT
GCTGCTGCTGCTGCCGCCGCTGGCTCTCCAAGCTCCAGCAGCAGCGGCCCCAGGGAGCAG
CCGCAGCCCGCCTCTCCTGCTCCACCCCCAGCAGCAGCAACAGCAGCAGCAGCAGCAGCA
GCAGCAGCAAAGGAGGCCTGGATCCACCCCAGCGGCCGCTTCGGATGTTCTCCTGCTGCT
GTTTGCATTTTAGAAAGAAGTCCCCACGAGCCTCCGGTGCTGGACTGTACCTGCAGCAGC
GGCACTCAGCAAAACCGCTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00000920 | 94 S | GSSSSKKRL | 0.993 | unsp | ETH_00000920 | 228 S | ASLESREGQ | 0.994 | unsp |


