

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00001425 | OTHER | 0.999302 | 0.000149 | 0.000549 |
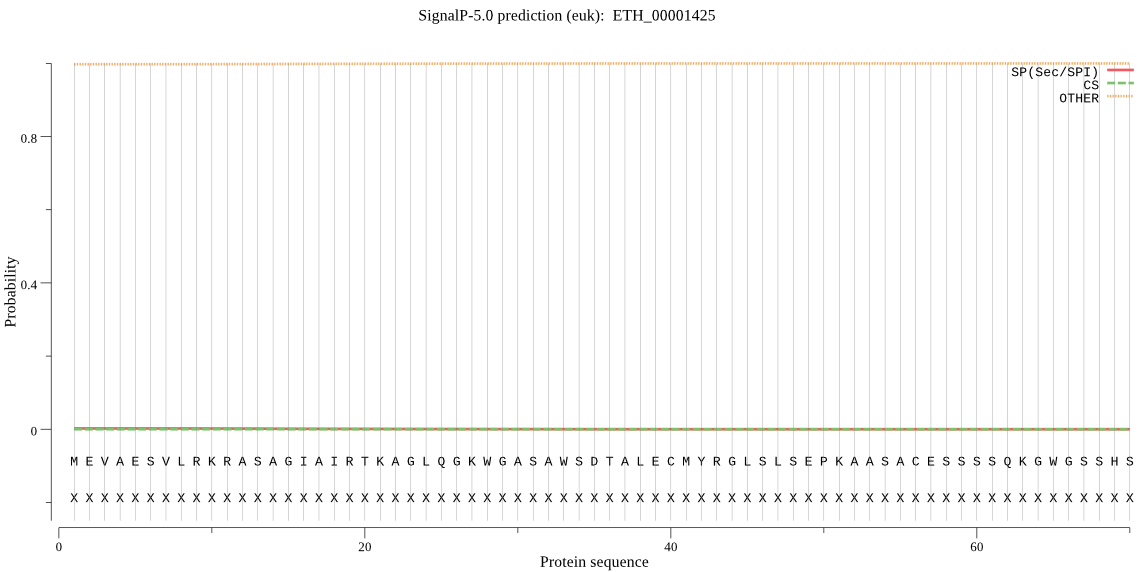
MEVAESVLRKRASAGIAIRTKAGLQGKWGASAWSDTALECMYRGLSLSEPKAASACESSS SQKGWGSSHSYPFLPAPRDIFGGTRSSVRGDTEVKDEWLLASPVRDVGTALARSSGKKQQ HPESEDDNEAAQQLRSQLLQLRAEGHRRAARLRRERQLLQQQLAEARQQQQVLLLRQQQR SICTSVGASQGSCDPWGPPTLSVPPPAPAVVRRGPRGGPLPPFSPRMVLLHVYDLTPAIS NYVNRIMRPLGAGAFHAGVEVYGQEYSFGQTTATAGALRHYGSATASLEPSTLWTAAATL PGAAVVLVILDTDETTGVSVCQPMQHPAHVYRETVPMGPADLGMDEFLSLVEVLKVEWPG NSYDVLSRNCVNFADHLCVLLGVGHVPPWLFRLQRQANSIKGGARAAAAAAAAAARQLQR LNTQARISATAMAAAEAAATAAAVAAGAAVSFGRFIKHAHQEVRLSEEMGRLLVEGMSFL SERFSEGVSCIPSEWNEEETTALEPLHLFDRAPTPPEGLIAESCGATSEDSAEKLSESPV EKPLEKQGDERLEELRTSSLVNPEEGTPRAPRGATEESLQGATILKSWSQGSLSSLSQDT EQQRKSPSDWPYAADAILP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00001425 | 115 S | LARSSGKKQ | 0.995 | unsp | ETH_00001425 | 115 S | LARSSGKKQ | 0.995 | unsp | ETH_00001425 | 115 S | LARSSGKKQ | 0.995 | unsp | ETH_00001425 | 124 S | QHPESEDDN | 0.997 | unsp | ETH_00001425 | 466 S | EVRLSEEMG | 0.997 | unsp | ETH_00001425 | 538 S | KLSESPVEK | 0.995 | unsp | ETH_00001425 | 567 T | PEEGTPRAP | 0.992 | unsp | ETH_00001425 | 606 S | QQRKSPSDW | 0.998 | unsp | ETH_00001425 | 46 S | YRGLSLSEP | 0.997 | unsp | ETH_00001425 | 87 S | GTRSSVRGD | 0.994 | unsp |