

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
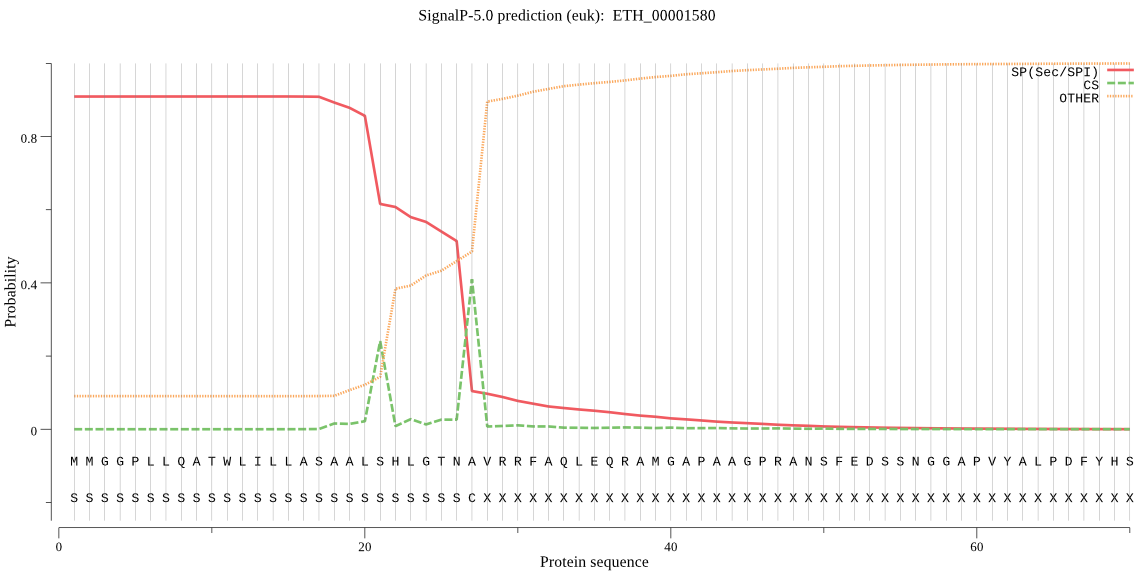
| ETH_00001580 | SP | 0.013766 | 0.966915 | 0.019319 | CS pos: 27-28. TNA-VR. Pr: 0.4214 |
MMGGPLLQATWLILLASAALSHLGTNAVRRFAQLEQRAMGAPAAGPRANSFEDSSNGGAP VYALPDFYHSYASMKKELRDLGKSCGLSVHQAALGGVTVEYFHLKESPDPSERLFLLPGD HPRELVAPEVVFSFLKALCSSQEAPEGYSAEALRQKLHVMAVLAVSVPARRAVEKGAYCM RSSPKGVDFNRNWGQSGDSAAAAEKSHPKYPRAPQQPEIQILKSLLKRFQPTVFVSVHSG RFGVYTSNEAPSGEPEKPPSTPSGGAGPPDHLQEAAEGAPTTAGESREQQIAGVVHASAC SGHSFDCEFGSIDMFFEAAGNEIDFVKAHLTTDEGEPPVALALEVYRQGNLSNEDAKLKE AKAQLLSLLKAPEELLDLTLPAAADVADLLPQETEGEDSLYTAEALIKWQLTRGPLDRLS CFLSFNPSTQSALNSVVRLWVEALFALAESL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00001580 | 252 S | NEAPSGEPE | 0.995 | unsp | ETH_00001580 | 252 S | NEAPSGEPE | 0.995 | unsp | ETH_00001580 | 252 S | NEAPSGEPE | 0.995 | unsp | ETH_00001580 | 286 S | TAGESREQQ | 0.991 | unsp | ETH_00001580 | 352 S | QGNLSNEDA | 0.998 | unsp | ETH_00001580 | 50 S | PRANSFEDS | 0.998 | unsp | ETH_00001580 | 183 S | CMRSSPKGV | 0.995 | unsp |