

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00001730 | SP | 0.006654 | 0.993006 | 0.000339 | CS pos: 20-21. ALA-ND. Pr: 0.9081 |
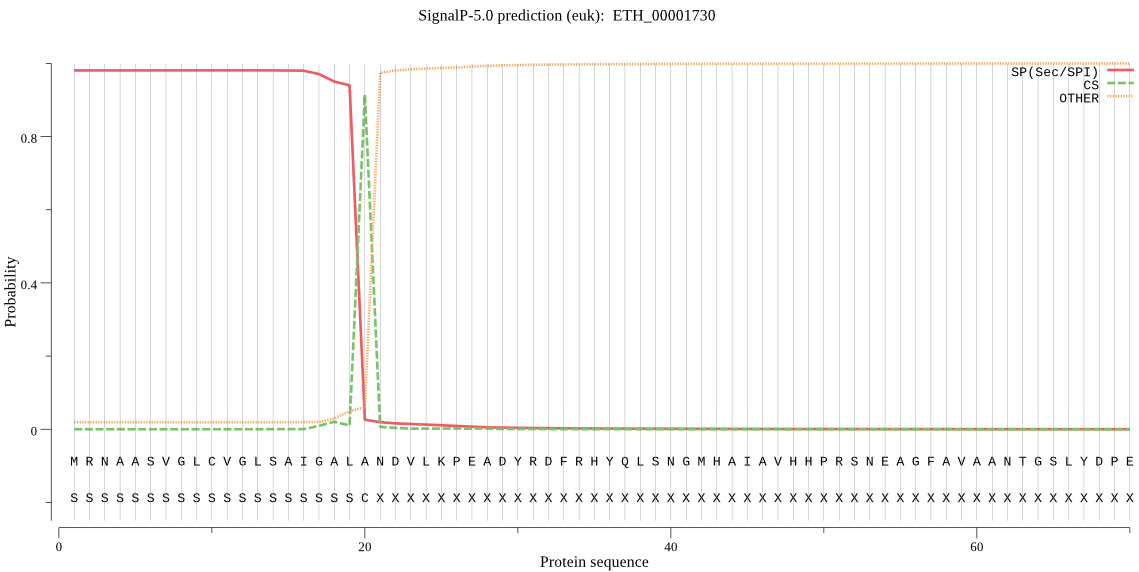
MRNAASVGLCVGLSAIGALANDVLKPEADYRDFRHYQLSNGMHAIAVHHPRSNEAGFAVA ANTGSLYDPEDVPGLAHFLEHMLFLGTSKYPEPESYDSFLTQNGGANNAYTDEEKTVFFN KVTDSAFEEALDRFSEFFKAPLFDRKYEEKEVNAIDAEHQKNIPNDDERAWYTIRSLAKG PMSRFATGNSATLSTNPKAKGIDLVDRLRDFHSKYYCGSNMCPGPMFDTVKPFDHTNSGK FIHLQSFSSQPSLWVAFGLPPTLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGIS PVVDRNTISTLLGLKVDLTQKGATHRGLVLQEIFSYINFLRDHGVGHDLVSTLAQQSHVD FHTTQPSSSLMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAF ADPEFSSKVESFETDPYYGVQFRVLDLPQHHAVAMTVLTASPNAFRMPPPLLHIPKASEL EILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQG SVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRDP SVVESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDD SIAKLSELKNVHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIE ATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAK ESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTK VEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDPSRRMVSLF DLLLNLPSYKPEKRQVVKLIADLEPAKEVTLIGEAKGQDAALLRRGDTAEEVVKAPTKLL SLEDKFVSQVQQANGYFPDVSICELPPSLLSLFELLEDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00001730 | 645 S | NDAFSRMDL | 0.996 | unsp | ETH_00001730 | 645 S | NDAFSRMDL | 0.996 | unsp | ETH_00001730 | 645 S | NDAFSRMDL | 0.996 | unsp | ETH_00001730 | 799 S | EGSKSHPDE | 0.993 | unsp | ETH_00001730 | 898 S | RRMVSLFDL | 0.991 | unsp | ETH_00001730 | 961 S | TKLLSLEDK | 0.996 | unsp | ETH_00001730 | 369 S | QPSSSLMDE | 0.996 | unsp | ETH_00001730 | 601 S | LRDPSVVES | 0.996 | unsp |