

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
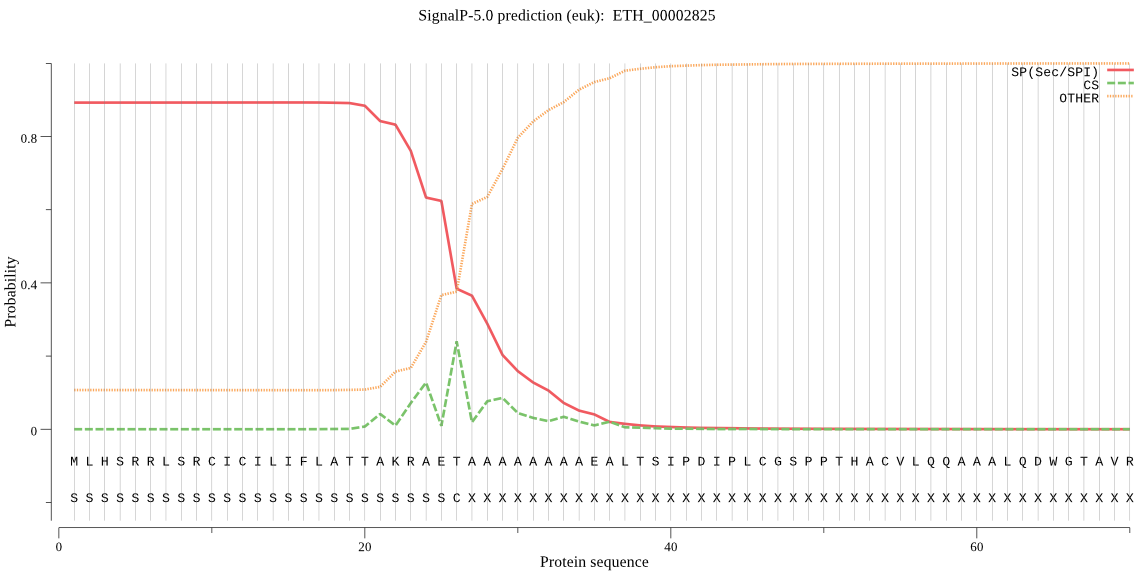
| ETH_00002825 | SP | 0.095376 | 0.731329 | 0.173295 | CS pos: 26-27. AET-AA. Pr: 0.4395 |
MLHSRRLSRCICILIFLATTAKRAETAAAAAAAAEALTSIPDIPLCGSPPTHACVLQQAA ALQDWGTAVRRQLHRHPELMYEELQTSILVQQVLTKLGIKHTTGWGRDRRTSLVDESLLE PSTERGDAEDLQKLQELQQQQQQQQQKEAGVVGTGVVAEIGTGEAPCVLLRADMDALPIV EQADVGFKSEVLGKMHACGHDVHTTMLLMAAAILKKNESSLKGTVRLVFQPAEEGGEGAR MMLEEGLLQQQPTASLALGLHVAPQLPTGTIASRAGNLMAATASFHFEVVGVGGHGAMPF ETHDPIAACAAAVQNINTFVARENDYSAESSGFVSVTQIQAGSAFNVIPSKCTLKGTIRA FSNEKLRELQLRLRQVVQHTAEAFRCSLRVIRLLTNTPALRVHPEALAVLHEAAPGIVAG GQVITMEPTYGGEDFAFVLDKLPGAFAFIGIGSGAEKAGHVPTAFGLHHPKFAADEAVLQ VGAALEAKFAFEAIKHLLQQQKPQGQHMPSQEPQQQKEEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00002825 | 220 S | KNESSLKGT | 0.993 | unsp | ETH_00002825 | 220 S | KNESSLKGT | 0.993 | unsp | ETH_00002825 | 220 S | KNESSLKGT | 0.993 | unsp | ETH_00002825 | 362 S | IRAFSNEKL | 0.996 | unsp | ETH_00002825 | 112 S | DRRTSLVDE | 0.998 | unsp | ETH_00002825 | 122 S | LLEPSTERG | 0.993 | unsp |