

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
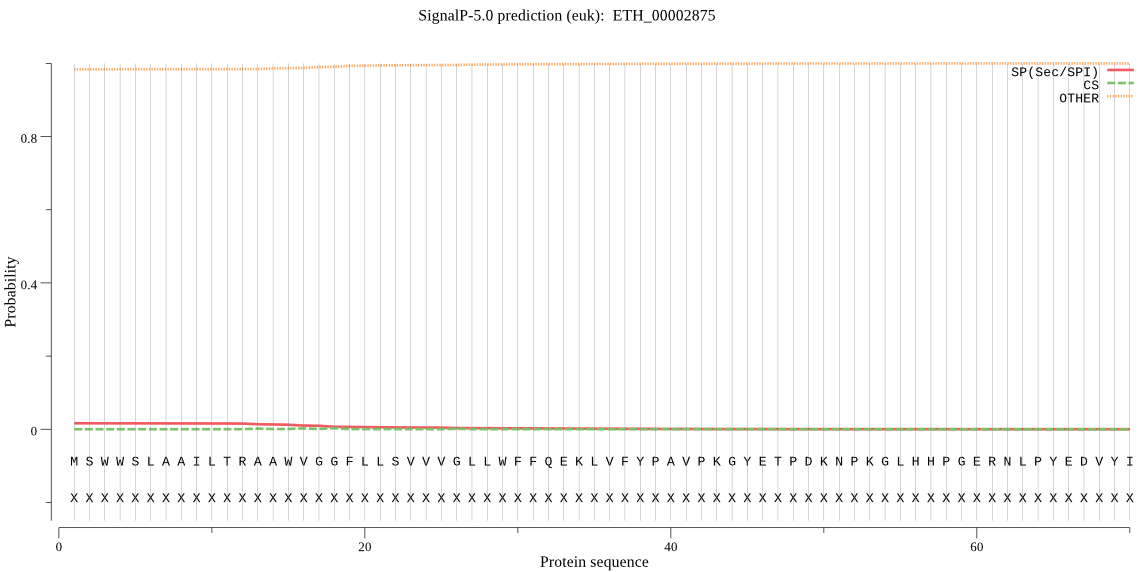
| ETH_00002875 | OTHER | 0.903375 | 0.089544 | 0.007081 |
MSWWSLAAILTRAAWVGGFLLSVVVGLLWFFQEKLVFYPAVPKGYETPDKNPKGLHHPGE RNLPYEDVYIRTSDGLTLHGWLLRQPNARQAPTFLYFHGNAGNIGFRLPNLELMFRMVGV NILIISYRGYGYSEGSPTEEGVYTDAEAALDFLLENKTVNNRDIFVFGRSIGGAVAIELA RRRSDELKGIVVENTFTSLLEMLYIVFPFLGPFNLLVKAVQRMYMLNDEKVRTLTLPVLF ISGRQDKLVPPSHMDVLYSSCGARLKLREDVENGGHNDTWEVGGQSYYNRLREFVQVALS ASTSSEVSRASGAHSTISDTSTDVSLRRLQCSSAAKEKAVCT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00002875 | 136 S | YSEGSPTEE | 0.995 | unsp | ETH_00002875 | 184 S | ARRRSDELK | 0.991 | unsp |