

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
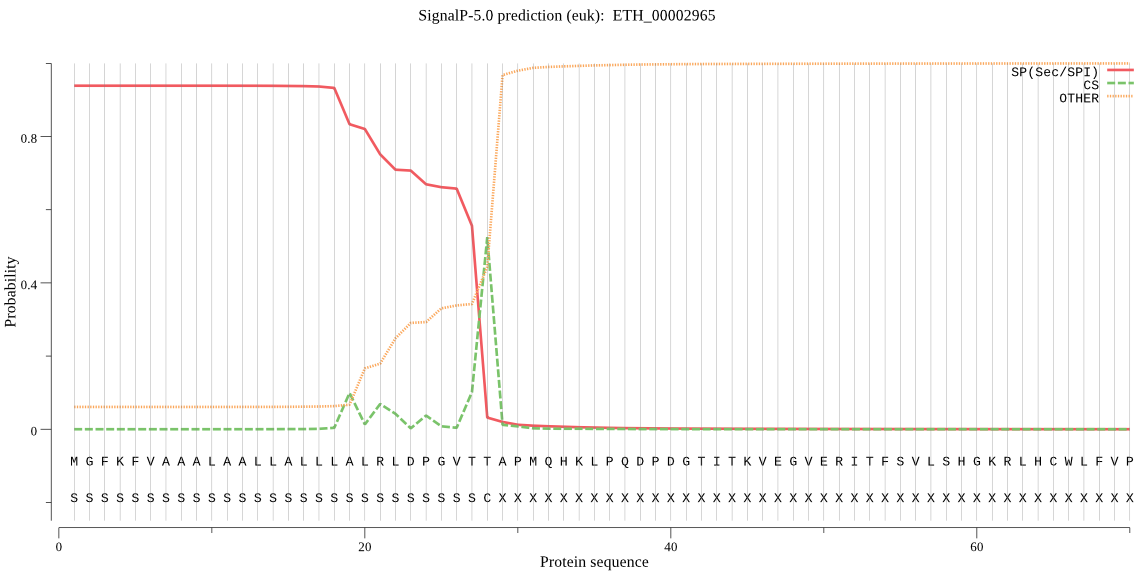
| ETH_00002965 | SP | 0.012541 | 0.987128 | 0.000331 | CS pos: 28-29. VTT-AP. Pr: 0.4247 |
MGFKFVAAALAALLALLLALRLDPGVTTAPMQHKLPQDPDGTITKVEGVERITFSVLSHG KRLHCWLFVPPQPLAKPPPAVVAAYGLGVQKDIAMVPLALALAREGFAAVLFDYRHWGVS EGLPRHAAVPQQQVEDLTALLQHLGAPGPLSSKLDSSKMALYGASLGGGVVLAAAAELCR RKDPLSSSIKAVVAAIPFVSGSKTSSKALQERGLLETARVGLAVVQDLVRSVFTAESAVY IPLARSSSSSSSSSSGISAMQLESNEHQIWASRTPLKGQVVDGAWENKLAARSLFWLSRF EVEDLAAEFLEIPTLLIAGSIDTVCPIEAIRDLVSRHQKKQSSSKMVLKEFPLRHFDFLT QQNFPILVSNTVQFIKEQI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00002965 | 156 S | SKLDSSKMA | 0.994 | unsp | ETH_00002965 | 250 S | SSSSSSSSS | 0.993 | unsp |