

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00003350 | OTHER | 0.781530 | 0.160892 | 0.057577 |
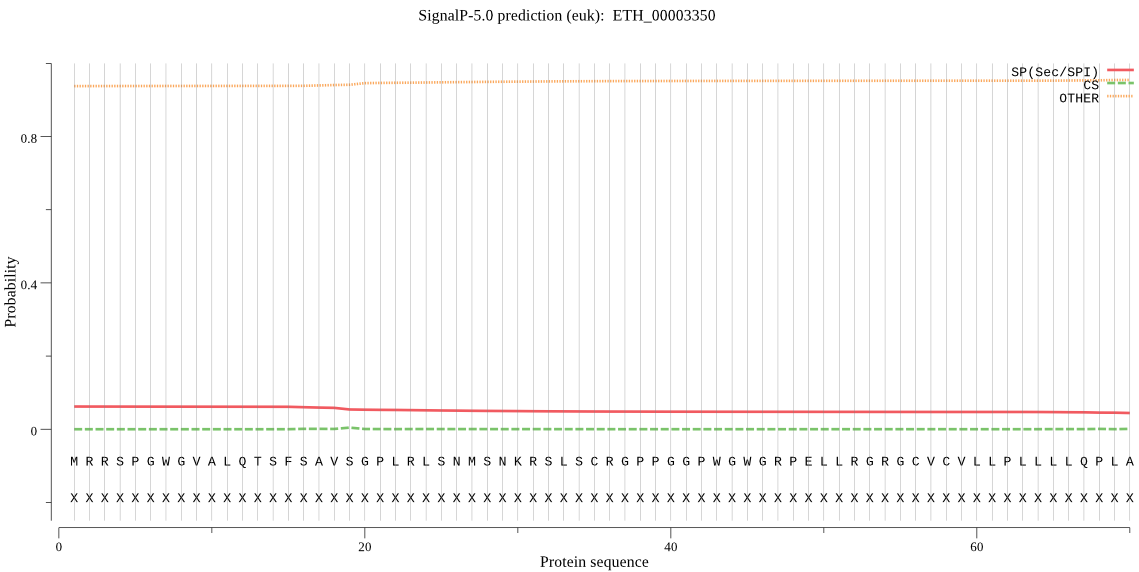
MRRSPGWGVALQTSFSAVSGPLRLSNMSNKRSLSCRGPPGGPWGWGRPELLRGRGCVCVL LPLLLLQPLAAAAAAAAGPAGAPGAPPASPRGPPPLKPGAVCKSTLDPSGNPLEEALARG PPPPAGAPRGAPPSHPAFAVASDRLVGELGLRVTQYLHKASGLEVWNLESEKEEEEMAFN ICFRTPVSDSTGAAHVLEHAVLNGSELFAPAAAAAAAAAAPGFLLLQQRSLNSYANAWTW PDRTCFPFASSNSKDFQNLLSFYLSCVFRPLALAQPHHLRQEGWRYEAVPRSSSSSSSSS GSSSGGGGEGEAAVDCETSPELCELAFGGVVYSEMKGVWGIAAAAEELQRLQQLFPALQT YAQVAGGRPPDIPLLQQGALAAFHRALYVPRNAWITFFGGPRRGPQGGPPLDFLDKFLTQ NKIYSPQPPQVVGVQHSFKAPVYAEFPFASLAKDPKDLISVSWVLNPCPAPPSGPPEGGP QGAPEAGECPGGPGGPGRVALQVLDQLLLGSPACLLRKALAQSGLGLAVYAPGLDLDLKY AQFTVGLKEVQQQEGAAAAVERLVLQTLEAIAAKGFSAAEIEAAFNKVDFSLRETPRKEL PRGLIFSQRMAQELNYNRDPLGALEFEQDIWRLRERLAAGEKVFENMVKELLLENPHRVT LRMKSSAEYAEQQAAEERAALTAAEKEMGLEGIRQVAREHKALKLLQVTPQTQPDCSSSG SSSSSSSCSSRTDAGK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00003350 | 89 S | APPASPRGP | 0.994 | unsp | ETH_00003350 | 89 S | APPASPRGP | 0.994 | unsp | ETH_00003350 | 89 S | APPASPRGP | 0.994 | unsp | ETH_00003350 | 293 S | VPRSSSSSS | 0.993 | unsp | ETH_00003350 | 296 S | SSSSSSSSS | 0.992 | unsp | ETH_00003350 | 297 S | SSSSSSSSG | 0.99 | unsp | ETH_00003350 | 298 S | SSSSSSSGS | 0.99 | unsp | ETH_00003350 | 300 S | SSSSSGSSS | 0.99 | unsp | ETH_00003350 | 304 S | SGSSSGGGG | 0.991 | unsp | ETH_00003350 | 591 S | KVDFSLRET | 0.995 | unsp | ETH_00003350 | 595 T | SLRETPRKE | 0.994 | unsp | ETH_00003350 | 665 S | LRMKSSAEY | 0.991 | unsp | ETH_00003350 | 726 S | SSSSSSCSS | 0.991 | unsp | ETH_00003350 | 730 S | SSCSSRTDA | 0.995 | unsp | ETH_00003350 | 28 S | LSNMSNKRS | 0.993 | unsp | ETH_00003350 | 34 S | KRSLSCRGP | 0.993 | unsp |