

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
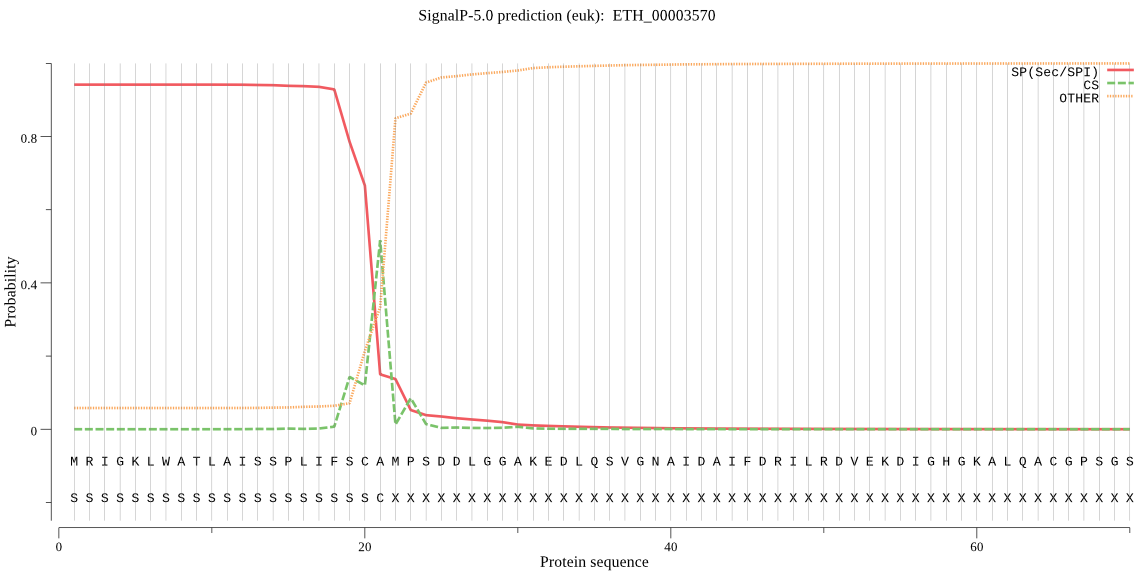
| ETH_00003570 | SP | 0.028253 | 0.968999 | 0.002748 | CS pos: 21-22. SCA-MP. Pr: 0.5788 |
MRIGKLWATLAISSPLIFSCAMPSDDLGGAKEDLQSVGNAIDAIFDRILRDVEKDIGHGK ALQACGPSGSHTQGVENCRRVLKGEPDESHELLPYLLKDQEAMRLWRDHFRRHVGELVSS ASEHSGADTPLQGPVLKSLAESEFWGSRPAVSNGALQHLRVKMQRLKLQAAEQGLDPEQA VTWEAEVSPRFKYHSIKDAKRHMGTYLSFYSDPDKPEVPLGEPLPVKVFAETQQVLETDK FDAREAFPQCAEVIGHVRDQGDCGSCWAFASTEALNDRFCIKSGGRHREALSPQHTTSCC DLLHCLSFGCSGGQPRMAWRWFSNDGVVTGGDYNELHTGKSCWPYEIPFCRHHSEGPYPK CEGPLPKAPKCRKDCEEAEYTSKVKPFKDDLHFATSAYSVEGRDQIKRELMENGTLTGAF LVYEDFLLYKEGVYHHVTGMPMGGHAVKVIGFGNEDGRDYWLAVNSWNEYWGDKGTFKIE MGEAGIDKEFCGGEPKVPNDKNASLLPSAQDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00003570 | 399 S | TSAYSVEGR | 0.991 | unsp | ETH_00003570 | 399 S | TSAYSVEGR | 0.991 | unsp | ETH_00003570 | 399 S | TSAYSVEGR | 0.991 | unsp | ETH_00003570 | 125 S | ASEHSGADT | 0.996 | unsp | ETH_00003570 | 195 S | FKYHSIKDA | 0.998 | unsp |