

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00004480 | OTHER | 0.999939 | 0.000059 | 0.000002 |
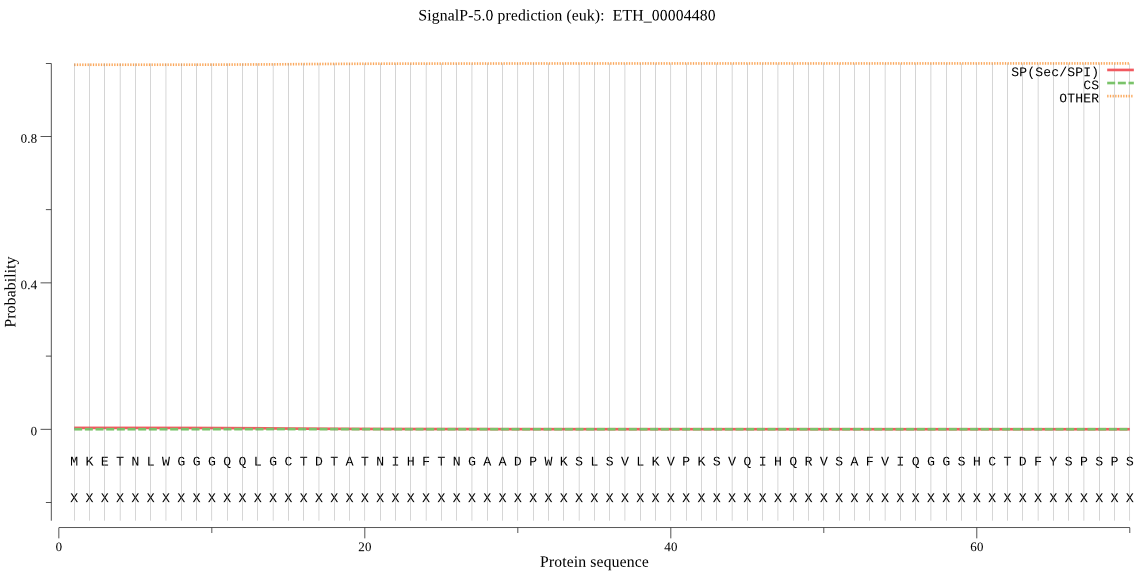
MKETNLWGGGQQLGCTDTATNIHFTNGAADPWKSLSVLKVPKSVQIHQRVSAFVIQGGSH CTDFYSPSPSDSSSLKEGRKAIKEAVASFIEAHRNPSSSSNSSSSSSSSRESGAAAAAAA AAVVAAAPAAANALAAAAAEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00004480 | 107 S | SSSSSSSRE | 0.993 | unsp | ETH_00004480 | 107 S | SSSSSSSRE | 0.993 | unsp | ETH_00004480 | 107 S | SSSSSSSRE | 0.993 | unsp | ETH_00004480 | 108 S | SSSSSSRES | 0.997 | unsp | ETH_00004480 | 109 S | SSSSSRESG | 0.996 | unsp | ETH_00004480 | 112 S | SSRESGAAA | 0.993 | unsp | ETH_00004480 | 74 S | SDSSSLKEG | 0.998 | unsp | ETH_00004480 | 105 S | NSSSSSSSS | 0.994 | unsp |