

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
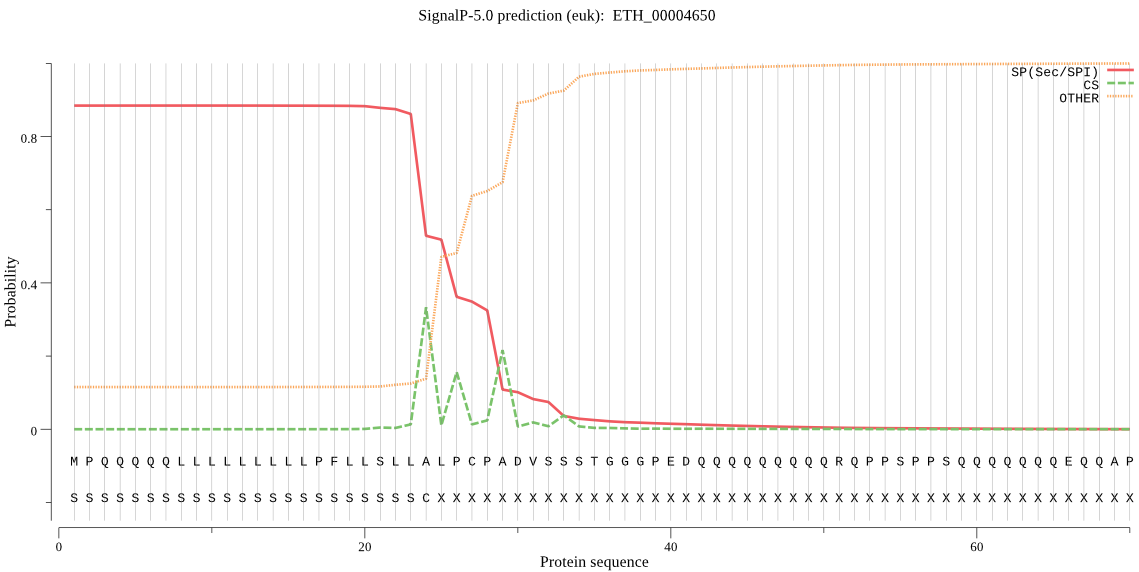
| ETH_00004650 | SP | 0.193573 | 0.806423 | 0.000005 | CS pos: 24-25. LLA-LP. Pr: 0.3201 |
MPQQQQQLLLLLLLLLPFLLSLLALPCPADVSSSTGGGPEDQQQQQQQQQRQPPSPPSQQ QQQQQEQQAPPQERRPSGASEGVHLPAAAAAAAAAASAAAAAAAAAQQQQQQLLREFVAL GDPADTHAVVASSSRQWLNYRHMANALAFYRSLKQQGIKDKNIILMLAGQDGPCSPRSAF PAGVYRHRSRAQNLFGGHDACAAALHRQQQQQQQQQQQEQQQQQEECALFGEGWEGAEVD YSGEAANADALLRVLLGVHTPETQQQQRLRSSSSSNVVVFLTGHGGNEFLKFSDWQTLQQ QQLAAAVQRMQQQRRFRRLLLIAETCQAATLLSGVRTPHVLRLSSSLKGESSYSYFSDSS MGTSLVDRWTHSVVNFLNRHANSHAAAAAAAARSSSSSSSSSSGGGGGSSMGVGRLLPSV AALLDSLDPNFLMSRCGVEEEPIPGSPTPLTQLPLEVFFSNRRKPVAYNFAYPAAAAAAA ADSSAAAAAAAAAAAAAAEPLHLWHSEALRFFPALNAPMQQQQQQQHQQQQQQQQHQQQQ HQQHQQQQQQRQQQPQRQLQQQHFEWVDPAVAAPLAAVLAGVACVATAASL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00004650 | 77 S | ERRPSGASE | 0.991 | unsp | ETH_00004650 | 398 S | SSSSSSSSS | 0.993 | unsp |