-
Computed_GO_Component_IDs: GO:0016020
-
Computed_GO_Components: membrane
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs: GO:0009405
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>ETH_00004860
MEALREGFGLRRLCCISAVAAFCLFGAKPSQAAASNGSQVASNPWGDSMQKFNIPYTHGS
GVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRLQQPTTDSSIWPGNYLEKVPTKGSPQDT
RPLGGGFAMWDTTPVKISPLTLSELEALAEQQRAKNDPTSPASEKLAKVTDGLGLCAWWA
WATYVPNGTTNLNDKYRYPFVWNEETKVCTLLGVSMQLLEGAGKYCSVGDASPVLTWYCF
YPEKTTRPVSYNSPYVREDHATACPEKAILGAHFGTWDGTTCQRMKAAKQITVPNPTECG
KAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATMWPVGAFSKDEPRTQGVGTNYANWY
TNGTCEMYDMVPTCFTLAPNQFSFTSLGSADPSTAELPPCTEASEGWEIYGYCECGDGHS
TPWKCENGQWIGGSDDCNCSSILPVALGVSFGLLVPIAALIAYFIYKRKKETSIAKNPEK
KKLLDEDEERDEEFLKVQEKRKHKQSDLAQEAEPSFWGETPQDHTNVVVDHNAHDAYY
- Download Fasta
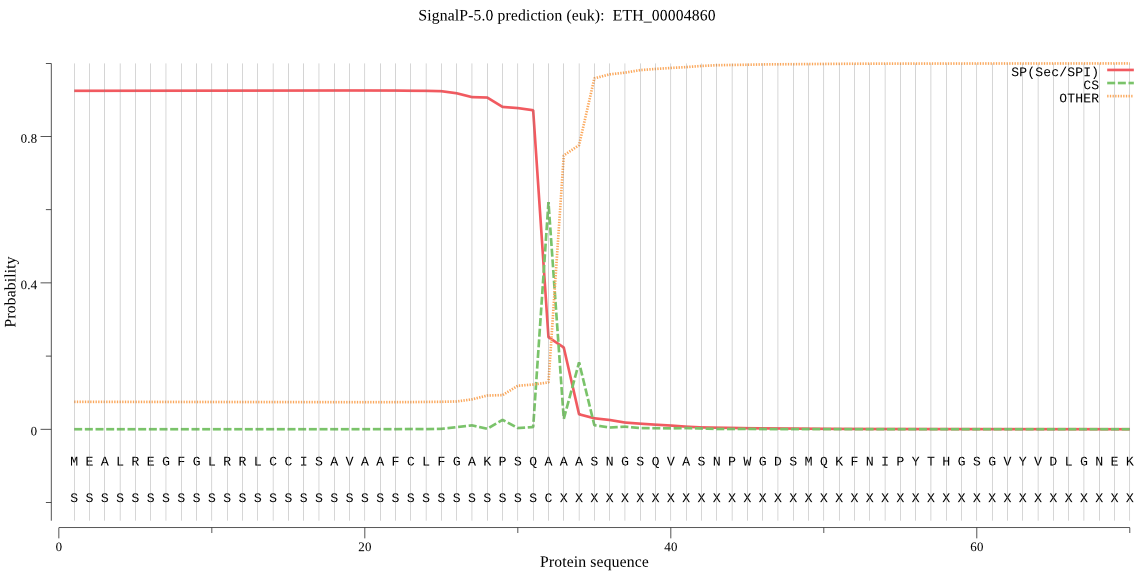
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00004860.fa
Sequence name : ETH_00004860
Sequence length : 538
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.609
CoefTot : 0.000
ChDiff : -11
ZoneTo : 1
KR : 0
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.124 2.459 0.449 0.861
MesoH : -0.125 0.284 -0.173 0.308
MuHd_075 : 33.643 17.188 9.308 6.203
MuHd_095 : 8.815 5.951 4.576 1.348
MuHd_100 : 20.512 9.290 6.563 3.680
MuHd_105 : 30.700 13.653 7.701 5.945
Hmax_075 : 16.217 16.683 4.557 5.588
Hmax_095 : 0.400 7.800 2.180 1.811
Hmax_100 : 2.600 8.800 2.340 2.670
Hmax_105 : 12.600 12.483 3.920 3.978
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9755 0.0245
DFMC : 0.9806 0.0194
-
Fasta :-
>ETH_00004860
ATGGAGGCTCTACGGGAAGGCTTCGGCCTGAGGCGGCTGTGCTGCATCAGTGCCGTGGCG
GCATTTTGCCTATTCGGAGCAAAGCCGAGCCAAGCAGCAGCTTCTAACGGCTCACAGGTA
GCTTCCAACCCATGGGGAGATTCCATGCAAAAGTTCAACATTCCATACACTCACGGTAGC
GGGGTGTACGTAGACCTCGGGAACGAAAAGACAGTGAGCAACAAGAAGTACCGCGAGCCT
GCGGGGCGGTGTCCTGTAATGGGTAAGGAGATACGGCTGCAGCAGCCCACGACGGACAGC
TCTATATGGCCTGGGAACTACCTGGAGAAGGTGCCCACAAAGGGGTCACCTCAGGACACG
CGGCCGCTGGGGGGTGGCTTCGCCATGTGGGATACGACGCCCGTCAAGATAAGCCCTTTG
ACTCTGTCCGAGTTGGAGGCGCTGGCGGAGCAGCAGCGGGCCAAAAACGATCCCACCTCC
CCTGCCTCGGAGAAGCTGGCGAAGGTCACTGACGGGCTGGGGCTGTGTGCGTGGTGGGCG
TGGGCGACGTACGTGCCCAACGGGACTACGAACTTGAACGACAAGTACAGATACCCTTTT
GTCTGGAACGAAGAAACCAAAGTGTGCACTTTGCTAGGAGTGTCCATGCAGCTGCTTGAG
GGCGCGGGCAAGTACTGCTCCGTCGGCGACGCCTCCCCAGTCCTCACTTGGTACTGCTTC
TACCCCGAAAAGACCACCCGGCCGGTTTCTTACAACTCTCCTTATGTGCGCGAAGACCAC
GCCACGGCTTGTCCCGAAAAAGCCATTTTAGGAGCTCATTTCGGGACTTGGGACGGGACC
ACTTGCCAGCGCATGAAGGCTGCGAAGCAAATAACCGTCCCCAACCCCACGGAGTGCGGC
AAAGCTGTCTTCAAAGTTTCTTCTTCAGACAACCCCACGCAGTACACGAAGCCACCCACC
ACTGAGGCCAGCAGCAGCACCAGCTCCAGCAACGCAGTGGCCACCATGTGGCCTGTGGGC
GCCTTTTCCAAGGACGAACCCCGCACGCAGGGGGTGGGCACCAACTATGCCAACTGGTAC
ACCAATGGTACTTGTGAGATGTATGACATGGTCCCCACTTGCTTCACCCTCGCCCCTAAC
CAGTTCTCGTTCACATCCCTGGGATCGGCCGATCCCAGCACCGCCGAGCTGCCCCCCTGC
ACCGAAGCCAGCGAGGGCTGGGAAATCTACGGCTACTGCGAGTGCGGAGACGGCCACTCG
ACGCCCTGGAAGTGTGAGAACGGCCAGTGGATTGGCGGCAGCGACGACTGCAACTGCAGC
AGCATCCTTCCCGTGGCCCTGGGCGTTAGCTTCGGCCTGCTTGTGCCGATTGCCGCTCTT
ATCGCCTACTTCATCTACAAAAGAAAAAAAGAAACCAGCATCGCAAAGAACCCTGAGAAG
AAAAAGCTGCTGGACGAAGACGAGGAGAGGGATGAGGAGTTCTTGAAAGTGCAGGAGAAG
AGGAAGCACAAACAAAGTGATTTGGCCCAAGAGGCCGAGCCTTCATTCTGGGGCGAAACT
CCCCAGGACCACACAAACGTTGTGGTCGACCACAACGCCCACGACGCCTACTACTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00004860 | 163 S | TSPASEKLA | 0.993 | unsp | ETH_00004860 | 163 S | TSPASEKLA | 0.993 | unsp | ETH_00004860 | 163 S | TSPASEKLA | 0.993 | unsp | ETH_00004860 | 245 T | YPEKTTRPV | 0.992 | unsp | ETH_00004860 | 250 S | TRPVSYNSP | 0.994 | unsp | ETH_00004860 | 307 S | VFKVSSSDN | 0.994 | unsp | ETH_00004860 | 328 S | SSSTSSSNA | 0.99 | unsp | ETH_00004860 | 343 S | VGAFSKDEP | 0.992 | unsp | ETH_00004860 | 116 S | PTKGSPQDT | 0.994 | unsp | ETH_00004860 | 160 S | NDPTSPASE | 0.995 | unsp |


