Fasta :-
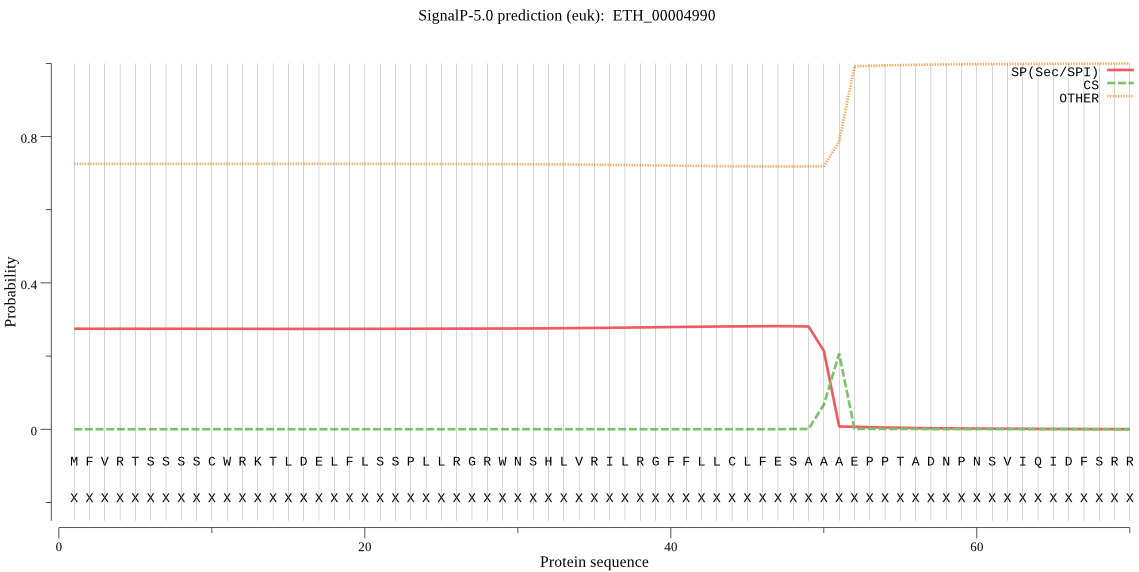
MFVRTSSSSCWRKTLDELFLSSPLLRGRWNSHLVRILRGFFLLCLFESAAAEPPTADNPN
SVIQIDFSRRLADVDIPECARNQIVYTTMADYEIASLLPQRSSSGLYMLDRLEHFLSETP
EADPDKVPLSGDSSQDATKTKCEMPTLLSHELHVGSPELQSYLRAARLRWLLDRCFRFSD
VGVDDWRYELCIGRKVTLFKSSALTGKTVGKNLFELGTYASSMDQLWADGTMTQWYTGGT
EGRQTEVRFVCGDTHPRVKGISGPQNGKYRVWLEAPMFCDYRENAREPALTESLIRSLEG
WCSTFTTDGWWSYEYCHPDSLVEYHKEPDGEVKGPLHLLGTLHASGDAAELIFKRPDPAN
PTLRGTDTLPPRANGAPRFKFLPVEIIDKPKQFSKSPTPASSKVLALQLTNGTVCEGTDK
QRSARVLFECPVDFPVLATHRIVQIEETSFCTYDLLIHTPLVCPHPKLLPPRPLDAQAVL
GYPVRRTFTYTITPASWKGLDAVDNAEETMGQTPTKEATNEQPVEAHLEALPKEAVEVLA
EGRNLDSFGLAKYIRKRVSVEEPKFKVGEIVRHRWWQYTAVVLSWDTSCMANNDWSSKAG
LEAVSKNISEVSHPDINKYFKVGQCLPVQCGPYEKLRRRQRIEDGAFHF