

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
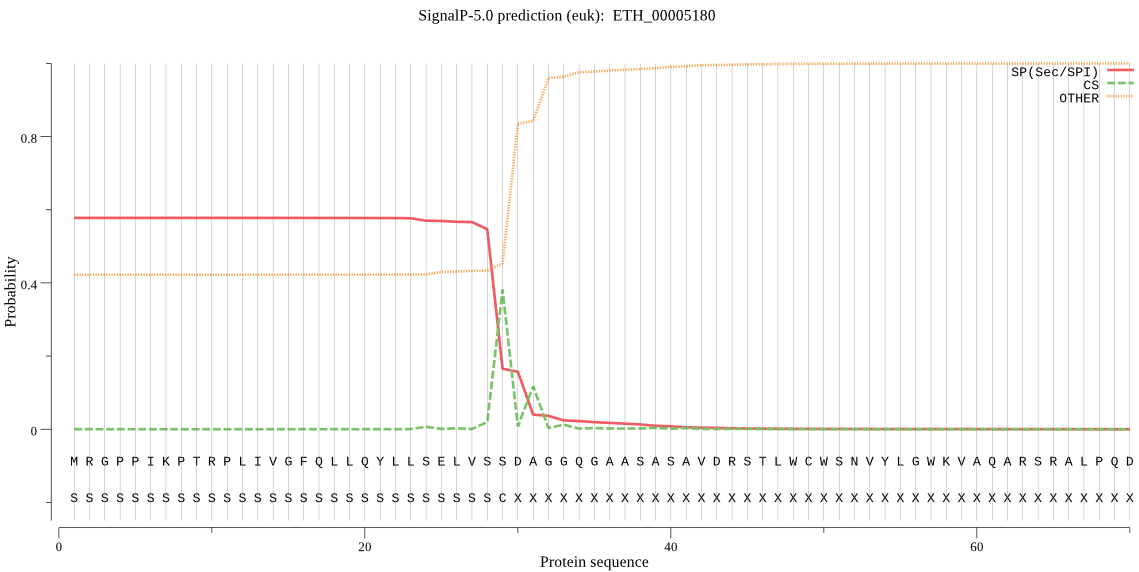
| ETH_00005180 | SP | 0.258085 | 0.732809 | 0.009106 | CS pos: 29-30. VSS-DA. Pr: 0.7630 |
MRGPPIKPTRPLIVGFQLLQYLLSELVSSDAGGQGAASASAVDRSTLWCWSNVYLGWKVA QARSRALPQDKQAAYWEQRHEHFANLVWDNIKELKGWWVKVGQFLSTRSDLLPQQYIHHL IKLQDMMPTTPFEVIEKTLRTELGDLNLIFSKIDEQPLASASIGQVHRAWLLDGTAVVVK VQHADVESLLMHDMANLKQLSWAFGMLEQGMNLTPILEEWQKAASKELDFRFEYAHQQRA FESAERSGIGVIIPKCYSNLVTKSVMVMEYIEGFKVTDTAKLDLYGVDRRELMYKLCDSF AYQIHIDGLFNGDPHPGNILVSINPVTKEAKPVILDWGLVKEFDSKGQTAFSKLVYSVAS MDVMGLMEAFEHMGFQFKEKKGAVVDPEIYMEALRVAFRQGEVEKKETEELHKAAGETVQ AAVKAGINRKKIQEANPLEDWPKDIIFFVRVASLLHGLCVQLKVHIPFLEIMVRRAQECL FNRYQPPSPLIYNYFLGGRGAAKCSLQQRLNRLLRRLVEQKLVLGCQVAVCFQGEMIIDA AIGQMGPVDTRPVTRDSLFCGYCTTKGILSTALLQLVSSGELELDDAVSSWWDGFIRYGK RGITVQQLLTHQAGLHRALPADLTLSRLADYSEMVRIVEDASPVCTPGSCGRYAYLTYGW AVSELVRCVSGVSIDVFIRERLLEPLGLEDEIFMRLPQEVEQTSSADQPSQLSACVLLES KKATSSALLENKATPLLNSSTPCSPSLRQAASDNSRTVPSTQCALNVTSSLELHPSSHSG EEKPPLAERMATVYSSSGSAGTNSGLPRTFQSANSPLAYPSSLSGGPQQFSPTELSPSFL LPGSQTPRIDTPEAGSSNREVQDHEDSRSRHNSNNSLRSLRSPLSDSDRARQRTENNSDF SVFSRPLESRVIGETDSSRRMLQVDDAASDRSKCSSTTFTHGTQNSEKVHQLSAFPKEVE IVTADTPFFWRLTSARRKISFSPMTQEEVLQRLKQTKGRLESTKGDGVLSSKDSNTRRSI ASAMRQRRCRLRNMKGNSGPLEQIREVKICSTRSAESLPDVFSRARAARHAEEERQKSEE EGALSHSDALTEGLRTPESDSDSACITPAAPGSAPRVSVLELIKTKPHVMDPLIYDSLRI VNQLIPPTNCRCTARALAKLYAALGEGRIISKPLLDAARVAHTVDPTLEALLLTGGGSRT WGLGYQLYECLYLSPPGWGGMRLGHGSQAGATLLHSRKPSTAEYVSRRSRRSRDSNIFAS FGSNAGPVFQNDVEALLRDRGLSSSHPWGATLGLKPRGVVGFGHGDVGGSVALCFPEMNL SISILLNDVLTGPEASQLILDFLLMCFGLQPKWQTQVSIDELLENLTPPLSQS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00005180 | 704 S | VEQTSSADQ | 0.993 | unsp | ETH_00005180 | 704 S | VEQTSSADQ | 0.993 | unsp | ETH_00005180 | 704 S | VEQTSSADQ | 0.993 | unsp | ETH_00005180 | 779 S | PSSHSGEEK | 0.996 | unsp | ETH_00005180 | 1002 S | GRLESTKGD | 0.994 | unsp | ETH_00005180 | 1010 S | DGVLSSKDS | 0.996 | unsp | ETH_00005180 | 1057 S | RSAESLPDV | 0.994 | unsp | ETH_00005180 | 1078 S | ERQKSEEEG | 0.998 | unsp | ETH_00005180 | 1085 S | EGALSHSDA | 0.996 | unsp | ETH_00005180 | 1099 S | RTPESDSDS | 0.994 | unsp | ETH_00005180 | 1118 S | APRVSVLEL | 0.991 | unsp | ETH_00005180 | 1249 S | VSRRSRRSR | 0.996 | unsp | ETH_00005180 | 1252 S | RSRRSRDSN | 0.997 | unsp | ETH_00005180 | 1255 S | RSRDSNIFA | 0.995 | unsp | ETH_00005180 | 590 S | DAVSSWWDG | 0.991 | unsp | ETH_00005180 | 642 S | VEDASPVCT | 0.993 | unsp |