

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
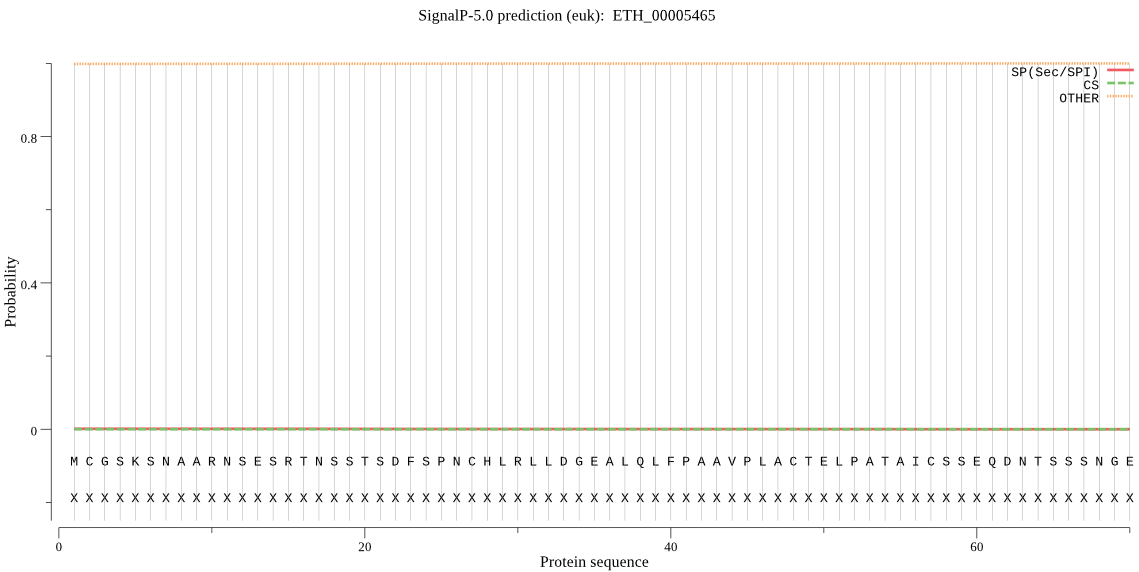
| ETH_00005465 | OTHER | 0.999992 | 0.000007 | 0.000001 |
MCGSKSNAARNSESRTNSSTSDFSPNCHLRLLDGEALQLFPAAVPLACTELPATAICSSE QDNTSSSNGEHETWYMVLHGILGGRRNLRSLAANLICGNTETTAQSSSSCSHQRGALRGR QRAFCFDLRGHGCSGRGELKLHLMARDVYAQIYLLLKELAKKRRVDMGSLLAEADSGANS KSGSDSGVLLPHWPGIIRVHALSKALDVRLVLVGHSFGGMVCMQSALDTLGLPVFAAVVV LDIAPSPYLLKPHLLSPEVASRESTSIFIPFDTNSSAKSSSSKRSSSSETGNSSSRIACC LDVARLVPPPKEGGYSSAALVSILADISLPLFSTKAAVTKVLQQLQPPINPAVINWLLLS LSTNKQRTAELEHLTRQQTCVSDRARIKQGVVMPHEMQRQQDMRQLQQHLVGQQFDDALC WHLDLLALQELIAANISMGLAAAPLSVHNPELTAAPGTATATGTSAAGSRPSYTGPCLFV RGALSPWLNYKQQQPSIHRHFPKATLREVTHAGHWLHAQQPQQTAATITQFLRALGLQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00005465 | 182 S | ANSKSGSDS | 0.993 | unsp | ETH_00005465 | 182 S | ANSKSGSDS | 0.993 | unsp | ETH_00005465 | 182 S | ANSKSGSDS | 0.993 | unsp | ETH_00005465 | 281 S | AKSSSSKRS | 0.997 | unsp | ETH_00005465 | 282 S | KSSSSKRSS | 0.995 | unsp | ETH_00005465 | 286 S | SKRSSSSET | 0.998 | unsp | ETH_00005465 | 287 S | KRSSSSETG | 0.995 | unsp | ETH_00005465 | 472 S | GSRPSYTGP | 0.991 | unsp | ETH_00005465 | 19 S | RTNSSTSDF | 0.994 | unsp | ETH_00005465 | 67 S | NTSSSNGEH | 0.995 | unsp |