-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
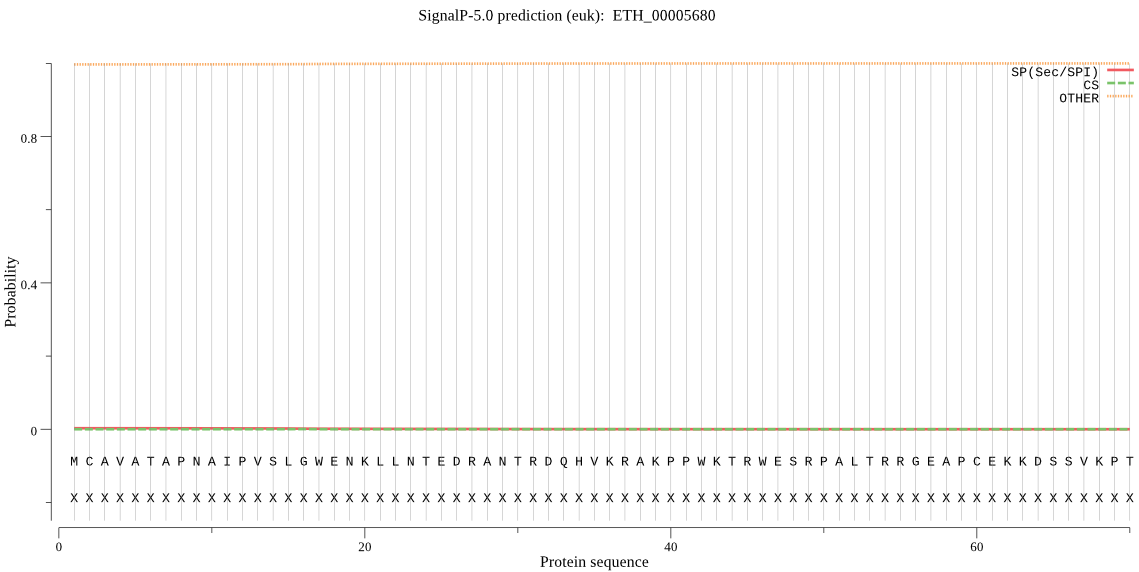
Fasta :-
>ETH_00005680
MCAVATAPNAIPVSLGWENKLLNTEDRANTRDQHVKRAKPPWKTRWESRPALTRRGEAPC
EKKDSSVKPTEVTGEGYRPEEAEIEPQWWRKTSAKEAYNKKRALFYVCATAVLRVEVAGS
PCGGALETGTSRSFISSKTVEWL
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00005680.fa
Sequence name : ETH_00005680
Sequence length : 143
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.857
CoefTot : -0.092
ChDiff : 6
ZoneTo : 17
KR : 0
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.318 1.559 0.141 0.508
MesoH : -1.526 -0.393 -0.613 -0.024
MuHd_075 : 18.410 10.036 5.877 2.784
MuHd_095 : 5.240 7.105 1.429 1.894
MuHd_100 : 1.584 6.092 0.658 1.503
MuHd_105 : 6.392 6.946 0.992 2.178
Hmax_075 : 15.983 15.283 2.544 5.052
Hmax_095 : 12.500 12.900 1.947 5.080
Hmax_100 : 13.100 13.000 1.984 5.100
Hmax_105 : 13.000 12.400 1.499 4.990
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9937 0.0063
DFMC : 0.9871 0.0129
-
Fasta :-
>ETH_00005680
ATGTGTGCAGTGGCGACTGCACCTAATGCCATTCCAGTCTCCCTCGGATGGGAAAACAAA
CTACTCAACACGGAGGATAGAGCTAATACAAGAGATCAGCATGTGAAAAGGGCTAAGCCG
CCGTGGAAAACGCGGTGGGAGTCACGACCTGCACTGACCCGGAGAGGAGAGGCTCCTTGC
GAGAAGAAGGACAGCAGCGTAAAGCCTACGGAGGTAACTGGAGAAGGCTACAGGCCAGAA
GAAGCAGAGATTGAGCCGCAGTGGTGGCGGAAGACTTCTGCGAAGGAGGCTTACAACAAA
AAGAGAGCACTGTTTTATGTGTGTGCGACAGCGGTACTCCGAGTTGAGGTGGCAGGGAGT
CCATGTGGAGGAGCTCTGGAGACAGGGACCTCGAGAAGTTTCATTAGTTCTAAAACAGTC
GAATGGCTCTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00005680 | 66 S | KKDSSVKPT | 0.998 | unsp | ETH_00005680 | 93 S | WRKTSAKEA | 0.994 | unsp |


