

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00005950 | OTHER | 0.999294 | 0.000089 | 0.000617 |
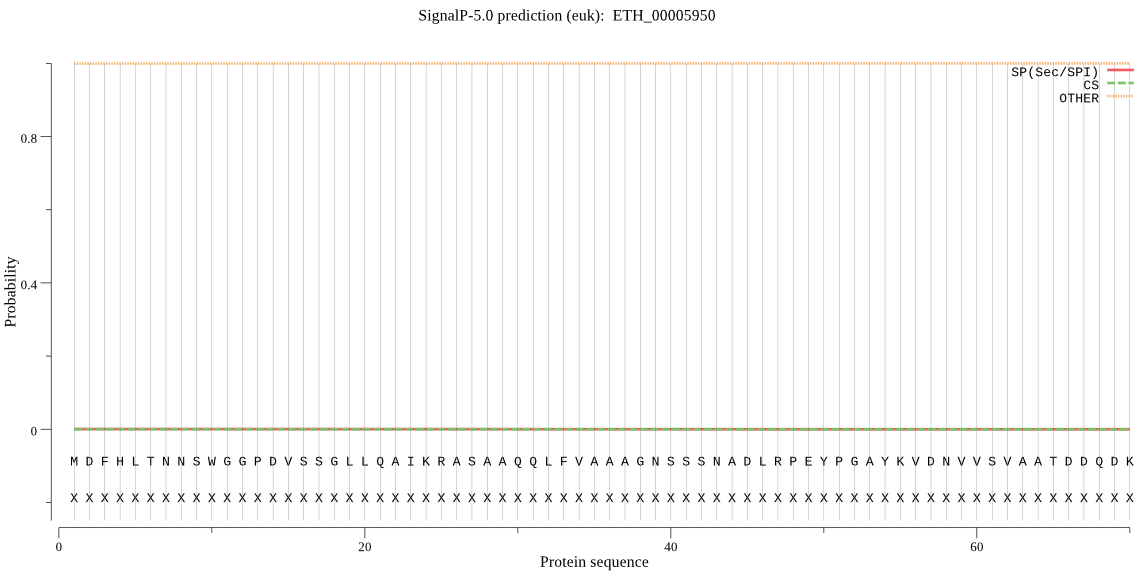
MDFHLTNNSWGGPDVSSGLLQAIKRASAAQQLFVAAAGNSSSNADLRPEYPGAYKVDNVV SVAATDDQDKLAAFSNFGKNAVHLAAPGVFIVSSFPPNTFKSLSGTSMAAPVVAGVAAML LSVRPMRVQELKDILFETADKPKSIRGEAQTLFRVSRALQPALFPFFALLCFAFFCFAFF CFALLCFLFFYKLRTNGRVSAGYAVCAAEAAAAAAAAAAAAERKNKSVILRKLKRQKEAA KKADSKLENKSENKSENKSENKSENKSEKAEEQEAPAARTRGCV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00005950 | 245 S | KKADSKLEN | 0.996 | unsp | ETH_00005950 | 245 S | KKADSKLEN | 0.996 | unsp | ETH_00005950 | 245 S | KKADSKLEN | 0.996 | unsp | ETH_00005950 | 267 S | SENKSEKAE | 0.997 | unsp | ETH_00005950 | 144 S | DKPKSIRGE | 0.993 | unsp | ETH_00005950 | 200 S | NGRVSAGYA | 0.993 | unsp |