-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
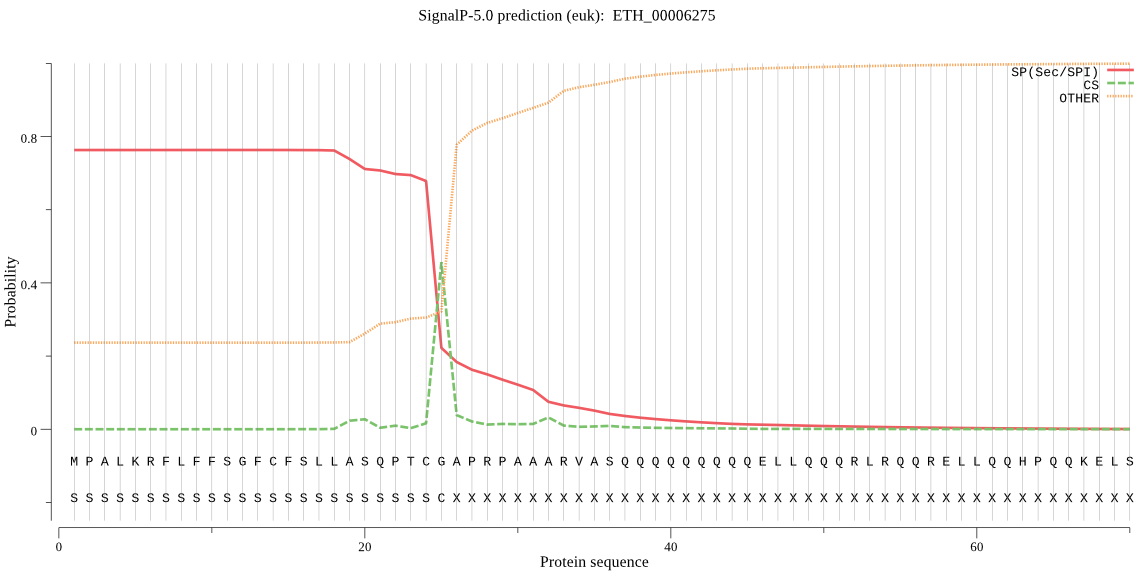
Fasta :-
>ETH_00006275
MPALKRFLFFSGFCFSLLASQPTCGAPRPAAARVASQQQQQQQQQELLQQQRLRQQRELL
QQHPQQKELSQLQQLQQEPLQQQHYQQDILQQELLQQELLAEQHQQQELLQQEQQEQEQE
QLVLLQQQQLQQQQLEQELQQQQQQQVEGEICSGEDVGFQGSGVRTPEVWGAAANEFPEV
LDLFEVKETEKEDSEEAKRVKETEKENETSFSKKEKMKENEMGAAAGLQFAEASRFGPLA
QYMQYLSTFTPGEQQQQQLQQLQQLQQLQQLLQQQQQQQLPQGAAAALKKLQRRAQAAAY
ANSVTSHPLLQGLIRSAVAAAAETLSSITLADLAAAATLPLPPIV
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00006275.fa
Sequence name : ETH_00006275
Sequence length : 345
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.727
CoefTot : -0.847
ChDiff : -16
ZoneTo : 45
KR : 4
DE : 0
CleavSite : 35
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.776 1.465 0.289 0.568
MesoH : -0.348 0.357 -0.333 0.179
MuHd_075 : 36.053 23.935 9.311 7.951
MuHd_095 : 22.825 15.894 6.619 5.558
MuHd_100 : 19.291 15.759 6.575 4.587
MuHd_105 : 21.590 18.661 7.047 4.911
Hmax_075 : 3.383 24.267 -1.009 1.692
Hmax_095 : 10.000 9.538 2.408 3.736
Hmax_100 : 19.800 18.700 4.410 6.140
Hmax_105 : 20.417 20.417 4.869 6.545
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.1442 0.8558
DFMC : 0.0565 0.9435
This protein is probably imported in mitochondria.
f(Ser) = 0.0889 f(Arg) = 0.0667 CMi = 0.54422
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>ETH_00006275
ATGCCGGCATTAAAACGATTCCTTTTCTTTTCGGGCTTTTGCTTCTCTCTTCTTGCCTCT
CAGCCCACTTGCGGAGCCCCCCGACCGGCAGCAGCTCGCGTAGCCTCGCAGCAGCAGCAG
CAGCAACAGCAGCAGGAGCTGCTGCAGCAGCAGCGGCTGCGACAGCAGCGCGAGTTACTG
CAGCAGCACCCGCAGCAGAAGGAGCTATCGCAGCTGCAGCAACTGCAGCAGGAGCCCCTA
CAGCAGCAGCATTACCAGCAAGACATTCTGCAGCAAGAGCTACTGCAGCAGGAGCTTCTG
GCGGAGCAGCATCAGCAGCAGGAGCTGCTGCAGCAGGAGCAGCAAGAGCAAGAGCAAGAG
CAGCTCGTGCTGCTGCAGCAGCAGCAACTGCAGCAGCAGCAACTGGAGCAGGAGCTTCAG
CAGCAGCAGCAGCAGCAGGTGGAGGGGGAGATTTGCTCGGGCGAGGATGTGGGGTTTCAA
GGTTCAGGTGTACGTACACCTGAAGTGTGGGGTGCTGCAGCAAATGAGTTTCCGGAAGTT
TTGGATTTATTTGAAGTAAAAGAAACAGAAAAAGAAGACTCGGAAGAAGCAAAAAGAGTA
AAAGAAACGGAAAAAGAAAACGAAACTTCTTTTTCAAAAAAAGAAAAAATGAAAGAAAAC
GAAATGGGCGCTGCAGCGGGACTCCAATTCGCTGAGGCCTCCCGCTTCGGGCCTTTGGCT
CAGTACATGCAGTACTTGTCAACGTTCACTCCTGGGGAGCAGCAGCAGCAGCAGCTGCAG
CAGCTGCAGCAGCTGCAGCAGCTGCAGCAGCTGCTGCAGCAGCAGCAGCAGCAGCAGCTG
CCCCAGGGAGCAGCAGCAGCACTCAAGAAACTCCAAAGAAGAGCACAAGCAGCAGCTTAT
GCAAACTCCGTCACTTCTCATCCGCTGCTGCAGGGCCTCATTCGTTCTGCTGTTGCTGCA
GCAGCAGAAACCCTCAGCAGCATCACTCTCGCTGATCTTGCTGCTGCAGCAACACTCCCC
CTGCCCCCAATCGTCTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
ETH_00006275 | 194 S | EKEDSEEAK | 0.997 | unsp | ETH_00006275 | 212 S | ETSFSKKEK | 0.99 | unsp |


