

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
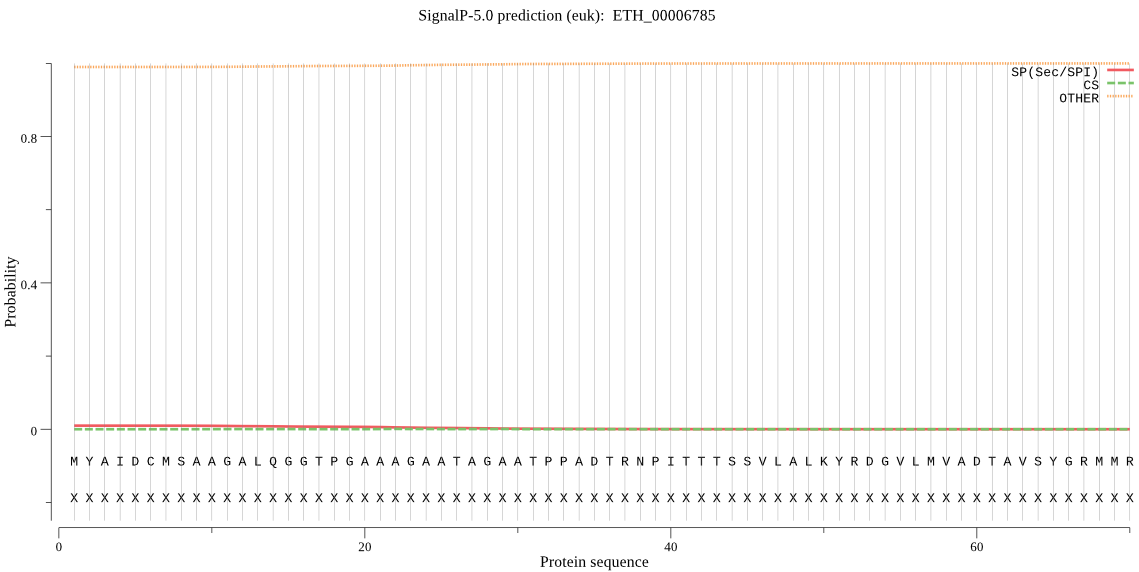
| ETH_00006785 | OTHER | 0.999772 | 0.000207 | 0.000021 |
MYAIDCMSAAGALQGGTPGAAAGAATAGAATPPADTRNPITTTSSVLALKYRDGVLMVAD TAVSYGRMMRFRGQPRFMNLDNKTLICTSGEYSDHQFLERELQELVDEDAAHADCSSSSS SSSSSTTLSPKAIASYTSRTMYQRRCKFNPYWLSVIVAGHMGSPKGIHEQQQQQQQQQQQ QEGQQQQGSEMYLGAIDMLGTFFEEQVVATGLGRYFAITLMRERQRPDMSEEEARELLEE CMRILFYRDCAASNEVQFAKVTASGVTVDEPKKLDSKWHFEAYTKKTIDMDMAGCSCTRL KKTKSSEEVYLLQAPAAAGAATAAAAAAAVAATAGAQAAARAMPPNGSCLG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00006785 | 230 S | RPDMSEEEA | 0.997 | unsp | ETH_00006785 | 230 S | RPDMSEEEA | 0.997 | unsp | ETH_00006785 | 230 S | RPDMSEEEA | 0.997 | unsp | ETH_00006785 | 305 S | KKTKSSEEV | 0.991 | unsp | ETH_00006785 | 122 S | SSSSSSSST | 0.993 | unsp | ETH_00006785 | 129 S | STTLSPKAI | 0.994 | unsp |