

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00007310 | OTHER | 0.999616 | 0.000141 | 0.000243 |
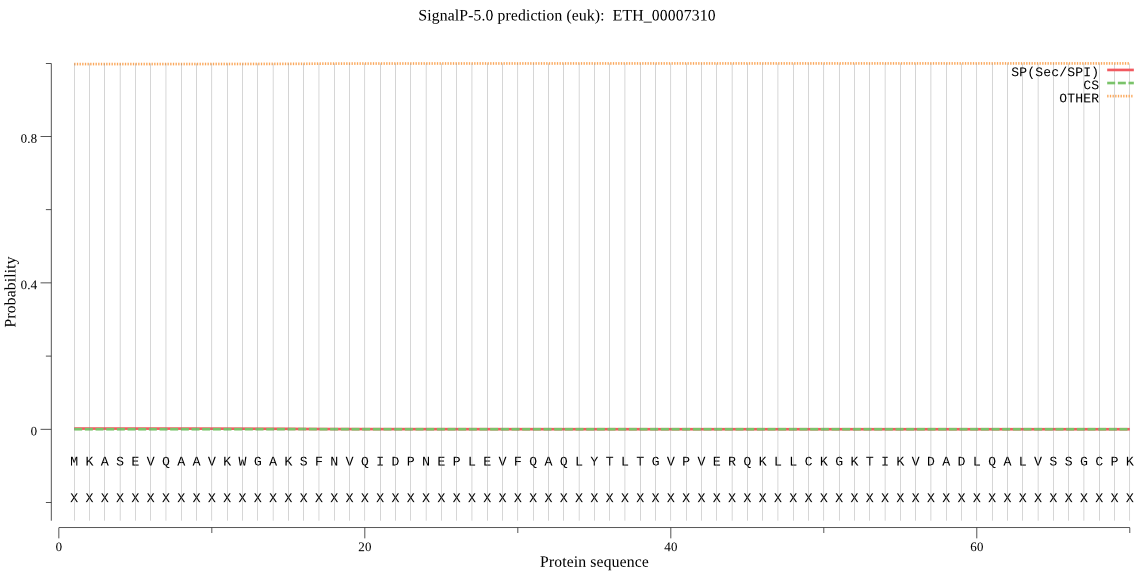
MKASEVQAAVKWGAKSFNVQIDPNEPLEVFQAQLYTLTGVPVERQKLLCKGKTIKVDADL QALVSSGCPKIMLMGTAEEKTAVKPPPEKTVFVEDLTPAQQAALLKEKKVEPLPNGIVNL GNTCYLASVLQMLRPAKDFSDLLKNYISGGAATAQFAATGQQGALRRLSLALKDFYNQWD QTVEAVSPFLLVPTLREAYPQFSRRSTSQAGTTGVPMQQDAEECLSCLMTAVAESLDSGA AAGLSKSLPYLPLKAGSSPLDLLFEVQVEVTSSRTKKEGEAEPKPTVQVERHRKLTCFLG TPQKPVSTIEDSLKFSLGDEVVNVGSTSAGASGTEEDTLIRSNRISSLALYLIVHFMRFE WKMGGSESEKAKICRSVKFSQTLDMFTYCTKELQDSFKVGRAIVALRREREASSGGAEKT TNGTASTESGAPAADGAAAATAAPEAAAVPAATNGSGAAHSAAAPAAEAAEAEAKKKETE LLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEP AAPSGPAAKRRKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLLMKHILVVPTEE ELLAVAKQL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00007310 | 316 S | SLKFSLGDE | 0.994 | unsp | ETH_00007310 | 316 S | SLKFSLGDE | 0.994 | unsp | ETH_00007310 | 316 S | SLKFSLGDE | 0.994 | unsp | ETH_00007310 | 332 S | SAGASGTEE | 0.996 | unsp | ETH_00007310 | 396 S | ELQDSFKVG | 0.991 | unsp | ETH_00007310 | 413 S | EREASSGGA | 0.995 | unsp | ETH_00007310 | 426 S | NGTASTESG | 0.994 | unsp | ETH_00007310 | 208 S | RRSTSQAGT | 0.994 | unsp | ETH_00007310 | 307 S | QKPVSTIED | 0.997 | unsp |