

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
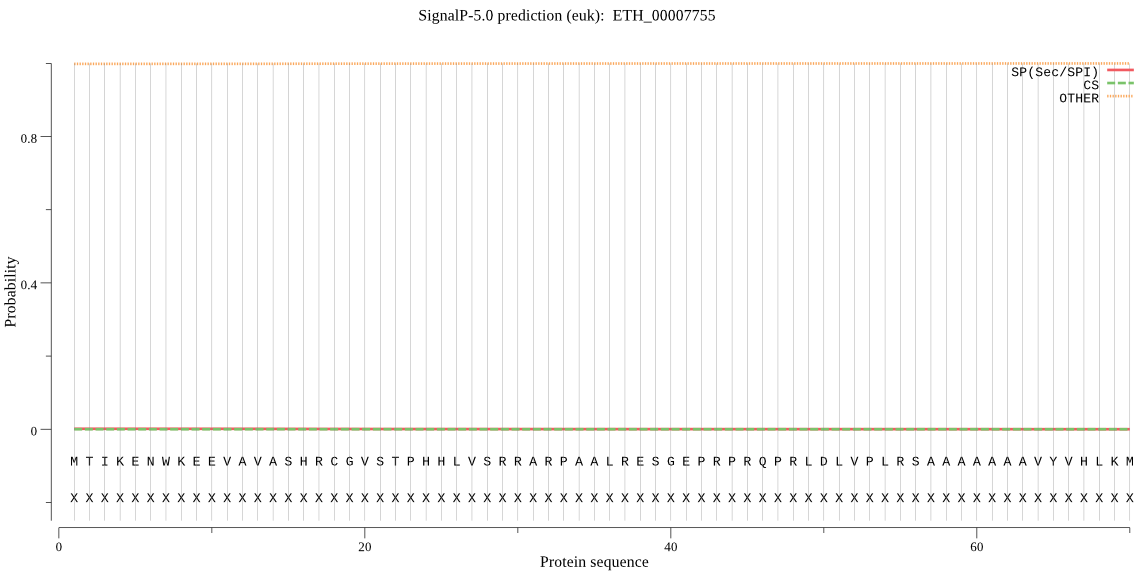
| ETH_00007755 | OTHER | 0.999914 | 0.000016 | 0.000069 |
MTIKENWKEEVAVASHRCGVSTPHHLVSRRARPAALRESGEPRPRQPRLDLVPLRSAAAA AAAVYVHLKMETDAAATQNQNNNTAAAGPDGSAAATQQPLKPGAESPAAAAAPAAAAPAA AASVPQGNGALPVKREEPLLSPKSEVDAAAAAAAAAPESAAAKPAAAGDAAAAPAAAAVK RDGDAAAAKGGSPPAASAAPAAAAAPAAAAAAPAAAAAAAAQLVALNRGEVELKLQRLFE MWEGRGINAKTEAWAAVDAICVFVGRAAEEDESAAERIQSWLTGFQFLETLFIFVRTTKE WLVLTSAKKAEHLRQLEAIGGLQIFLKDSQEGNAKSLQQMAAALRRAAGVDSDSGVRLAV FLGGSVLTTPGTLAADVIPWIKSFANKEENIEVPLSPLTATLTPSEMALVADASRVSVAM VKHQLVSRVEFVLDNGQNESHKSICDRAELLLKDQKQLQRLKEKCNLDPSEVEVLFKNVQ SGRHFDLRASALPSGEALSQSEGTIIISLGCKYKDFCAALCRTFILNGKKEQKETYELAL ELQQHLIGSLKVGVSFSAIHEKAVSFISSRKPELSRHLAKNVGHTIGAHYRDPRLVFNQK NDKVFVEPEMAFLVSVAFSDLKTAEGKEYAVWLADTVWLRPDGTTQVLTEGLSSQLAYVN YELDEGEEEAAESPLKAKSSKEGRREKREEGELVEKKKSREKGKEKEKEKEKKEKEKNSE KPEKPSSGTISASVLKNAESLILKDRLRRRDNGSAAHMEAAEERDNRQRELRKKKLSALQ QRFAREEAEDGGDEQQQKRQKKMQDLRAFSSPEEFPKDLRPSKLYVDMKSECLFVPFPGH HLPFHLSTVKNVTCSESEANLGAARAAAGSKPSRFSILRINFQVPGSQTLTQKGEENPLP DIAGKNQLFVKELMFKSEDSRHLQNIYRTIKEQLKRVKQKAAEDLQYPGELAAQEKLVLN RSGRRILLKDLMIRPNISTGMRKLIGALEAHTNGLRFTVNTRGQMDVVDITYSNIKHAIL QPCERELIVLVHFHLKAPILVGKKRTQDVQFYTEAGTQTDDLDNRRNRSYHDPDETLDEM REREMKKKLNGEFKRFVQQVEEVSKVEFDLPYRELRFSGVPLKSNVEILPTANCLVHLVE WPPFVLPLEDIEIVSFERVAHGLRNFDMDYNKPVKRIDLIPIEYLDNLKRWLNELEIVWY EGKQNLNWAAILKEIRDDPRGFVEDGGFDMFLGDVNGSEEEGEESEDDDDEEYAEQSSSD DDKEFNAEGSDSGESSGGGSDSDFSSDDSSLADESDDEEAAATDEDEEEGLSWDELEERA KKEDRKRRADEDSEGEETRTRKKRK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00007755 | 144 S | LSPKSEVDA | 0.993 | unsp | ETH_00007755 | 144 S | LSPKSEVDA | 0.993 | unsp | ETH_00007755 | 144 S | LSPKSEVDA | 0.993 | unsp | ETH_00007755 | 499 S | GEALSQSEG | 0.993 | unsp | ETH_00007755 | 679 S | LKAKSSKEG | 0.998 | unsp | ETH_00007755 | 699 S | EKKKSREKG | 0.996 | unsp | ETH_00007755 | 719 S | KEKNSEKPE | 0.995 | unsp | ETH_00007755 | 810 S | LRAFSSPEE | 0.994 | unsp | ETH_00007755 | 811 S | RAFSSPEEF | 0.993 | unsp | ETH_00007755 | 1069 S | RRNRSYHDP | 0.997 | unsp | ETH_00007755 | 1238 S | DVNGSEEEG | 0.997 | unsp | ETH_00007755 | 1245 S | EGEESEDDD | 0.996 | unsp | ETH_00007755 | 1257 S | YAEQSSSDD | 0.992 | unsp | ETH_00007755 | 1258 S | AEQSSSDDD | 0.991 | unsp | ETH_00007755 | 1259 S | EQSSSDDDK | 0.994 | unsp | ETH_00007755 | 1272 S | EGSDSGESS | 0.992 | unsp | ETH_00007755 | 1280 S | SGGGSDSDF | 0.993 | unsp | ETH_00007755 | 1285 S | DSDFSSDDS | 0.996 | unsp | ETH_00007755 | 1290 S | SDDSSLADE | 0.993 | unsp | ETH_00007755 | 1312 S | EEGLSWDEL | 0.994 | unsp | ETH_00007755 | 1333 S | ADEDSEGEE | 0.994 | unsp | ETH_00007755 | 39 S | ALRESGEPR | 0.991 | unsp | ETH_00007755 | 141 S | EPLLSPKSE | 0.994 | unsp |