

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00007910 | OTHER | 0.999962 | 0.000033 | 0.000005 |
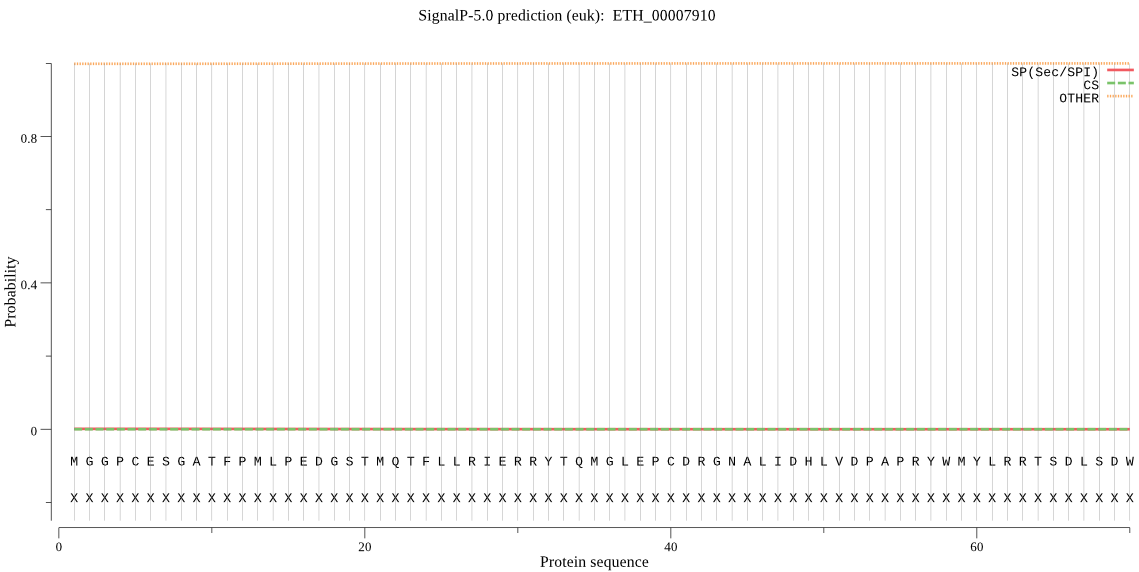
MGGPCESGATFPMLPEDGSTMQTFLLRIERRYTQMGLEPCDRGNALIDHLVDPAPRYWMY LRRTSDLSDWATVKRRLLERFDKTMSQSQFLTELAKVRWNGNPKEYTDQSAAVAERDVGI TPKELADYYCTGLPTDLLLLITGSKGERTKRPGETCKKCGSVGHYALDCPTYDRGRSGPE GKSSRQPTRLSTERTERLNDKVLLSRKHIPALKSCPKLEPRCRGPCGVIERIETIAYRLA LPVTYECHNVSHVSQLVPHCPHLPDLVSQGADAGWLPIRNAAGIPTKDYEENFIVDQRGP GADAPGPRQVARNP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00007910 | 177 S | DRGRSGPEG | 0.996 | unsp | ETH_00007910 | 191 S | PTRLSTERT | 0.998 | unsp |