

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00008785 | OTHER | 0.999782 | 0.000140 | 0.000078 |
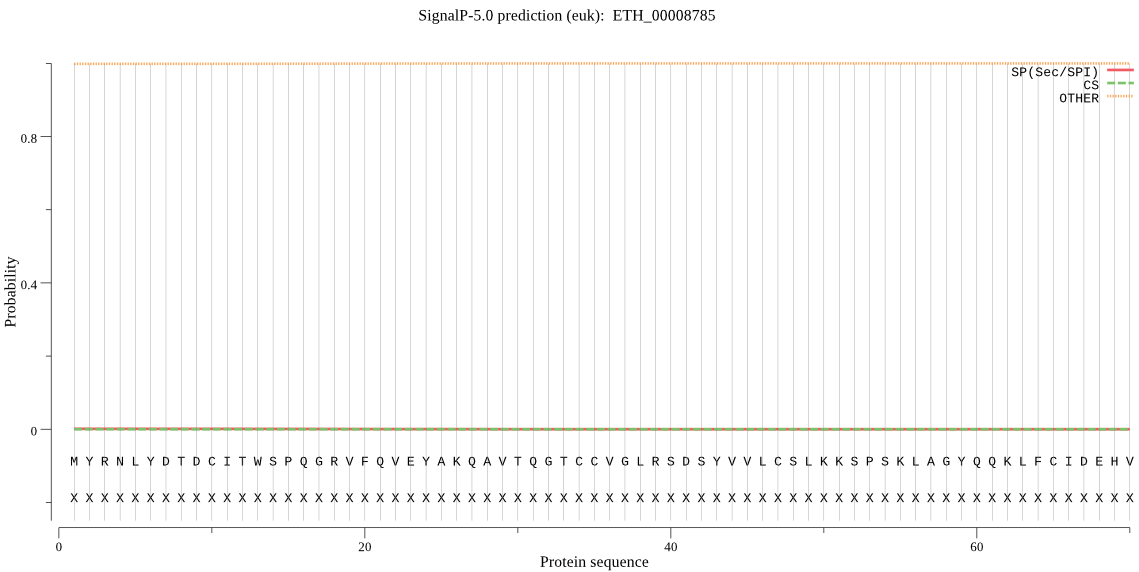
MYRNLYDTDCITWSPQGRVFQVEYAKQAVTQGTCCVGLRSDSYVVLCSLKKSPSKLAGYQ QKLFCIDEHVGVAISGITADAKVLCEIMRNACLQHKFTYDTEKPAARLALLVADKCQSNT QRSGKRPYGVGLLIAACDASGPRLFQNCPSGNYYEYNAVAFGARSQASKTYLEKHFETFP TGKLKTVNPSSLDDLLLHGAKALKASMAADTELTAECLCVGIVGRGQPWRELNHEELQTL VDRLTNAESSGDGGSMAVDMPE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00008785 | 123 S | NTQRSGKRP | 0.996 | unsp | ETH_00008785 | 191 S | VNPSSLDDL | 0.992 | unsp |