

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| ETH_00009260 | SP | 0.408996 | 0.589723 | 0.001281 | CS pos: 45-46. AWA-FL. Pr: 0.6768 |
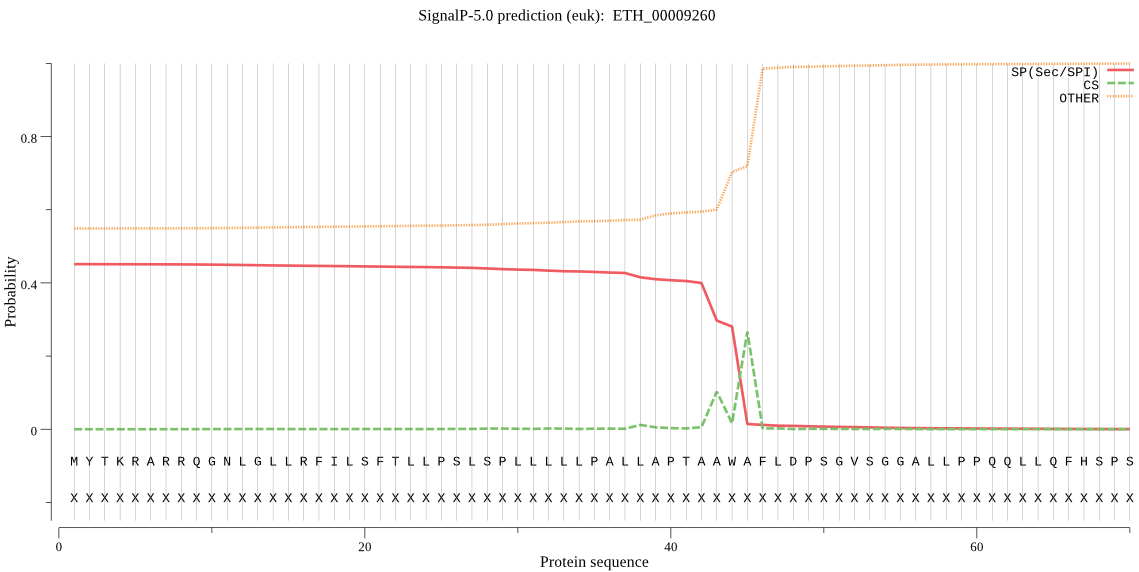
MYTKRARRQGNLGLLRFILSFTLLPSLSPLLLLLPALLAPTAAWAFLDPSGVSGGALLPP QQLLQFHSPSFSTPSLTPLSVIPSATSKQQLPAVGLSCRLSRAQPDGIECRELLRPLQTR PGRTASHIPQTAEAAAVKSATAAAAATANTQHMVPPTAPGLSSPLTGPQKAQVEEVARQS GPWMRDSDPLAAYVQMHGGKRPIRRILIANNGMAATKAIASMRQWTFETFGDANLLTFVV MATPEDLEANSEFLKKADAIVPVPGGPSRENYGNVSLICETAVKQKVDAVWPGWGHASEN PELPTRLAELGICFMGPNAKVMAALGDKIAANILAQTANVPSIPWSGDGLKAQLNEEGSI PEEVFKAATISTLEECRAAADRVGYPLLLKASGGGGGKGIRLCYKPEELESALSQVKAEV VGSPVFLMQLCERARHIEVQIVGDSFGNAVALSGRDCSTQRRFQKIFEEAPPTVVRPETF KEMERAAQRLTQSLGYEGVGTVEYLFNPATDSFYFLELNPRLQVEHPVTEAVTGQNLPAL QLQVAMGIPLNRIPSIRSFFGRDPTGTDKIDFLKEDYMPLKMHCIAARVTAENPSDGFRP TSGRVHSLSFNPPEKVWGYFCVGTKGALHAYADSQFGHVFAGGSTRNEARKRLIWGLQQL QIKGDIRTPINFLQVLLQQPAFVNHTLNTEWLDGIIKAKQLLPSPKADDVVFASAAFRAI QALEKQKASWLSTAARGHTYLPPVGPLLSVEVPIAWDGQLYKLNFTRTSPSALGFSINKQ KGEVQYRQQSDGSYLLSIYSDPPQEQQSDQQLQQMQQPQELQHGVHPRLLRFSATEEPLG LRLELGDNSLMVLDQRDPSEMRSDVNGRLVRYLVGDGEHVKKGQPFAEVEAMKMILTLTA GESGELIHRKSAGSLIQQGELLASLRLEDPSKVVRLTPFEGSLDLTAAAATPGEAEEETA ENTASPPASPSALQLSPEAAAKEVAAAAEARLELLLSGFVQQQTAQQLLNSLLSGPCGDS PLALAAFTEKCLTRFLETEELFAGRGNADAAAAVAAATTDTEGQFRVYVSHYALKQKTEM LLLLLKELAGRLTASGTSGFDSSRLEGLLRRVGSLRGVAYNVLALASQRLLKQLLLPSVA ERVETLRTQLQQLQNLQQQQQQQQLRDELLQLPEELGGYGLLAHLFRDSSVGLSAKEVML RRQLLGLEVQLQQQQQQGVIPFTWKDMLHEGPAATVNAGIFAAVSGMEDLQQQVGVLLQQ LGQQQKALSEETKGVLLLSLPVATAETAGAAEVADAAAAAAAAAAANSPDAAAALKRLHS LRLLLPRTNGDAQQVTLVPQQDLQLQEDPLQRGLSLSQFVFGEIEMMRGKGTVRRLPSAA AAAAVPPAVSTDLLLHAVSPKPPTPPTGAAAPLRTPALRETVYIHRIIPKADMLLQGEGL QQLLQHLLQQLEAAFRDPHVHPTSSSTVVLQLLQPEPEERQAALQKALFEAVHTLKAEQG ERVLRLNVEKVELRVPLAAGRATEAASAASAAASQQQQQIIRASSRGGCWLEPEELSVPG AVAEYDGECELHSSATATLRQQTQPKKQVLGAETSTADSSAAPSPLLSELSTPESASFKE GEQMASRSSAGDSMEFAASIQSSTCSSDRLPFSWGRGGTAAAAAEAERTAPLVPLKGVSE AQLAAKRAAAQRAGSTYIYDFLGFIKTELQQQWRRSQQKLENAGDSRKLKVPHKLMETRE FHMGPDGKLQLHRNWQVGDNKVGMVAFLLLLRTPEYPQGREVVVIGNDVTYQGGSFGVEE HEVYRQASAFARKHKIPRIYIACNSGARIGVYEQLKEKLHVEWTDPENPALGFKHLYITE KDMRELPPGAVVGHWIPTPAGAESDIGDRVFVLDAVVGSEGQHLGVENLRGSGKIAGETS LAYDEVFTLSLVSGRSVGIGAYVVRLAQRCIQQKTSPLVLTGYQALNKLLGREVYVSQDQ LGGPEVMLPNGIAHLEVANDKAGIAEVMRWLEFVPPTADATHTPHLETDPVNRDIEFTPT STPYDPRHMLAGHEVPATPPSSPLESDAEGAAEVPQQQHKKHQRRWVSGFCDRGSFKEVQ GPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEIPQAGQVWFPSSAY KTAQAIADFNRGENLPLIIFANWRGFSGGTRDMFNEVLKFGSMIVDALRVYKHPVFVYIP PHAELRGGSWVVIDPTINASQMELYADELARGGVLEPPGICEIKFREAERRKLMHKNDNT LKQWQADLATAATPKEAEKIKQNIKKREDQLMPVYTQVAHVYADLHDRAPRMAARGGVRR ILSWPKAREFFYWRVRRRLASNNAVKKLMAADPALTWDDALHLLTTEIPQTADSVDDKEA VAFFESSEGCEKLGALLRRTEAASSRRELLQLVSRLSAAEKKDVLSEVQSLLSK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETH_00009260 | 833 S | LLRFSATEE | 0.997 | unsp | ETH_00009260 | 833 S | LLRFSATEE | 0.997 | unsp | ETH_00009260 | 833 S | LLRFSATEE | 0.997 | unsp | ETH_00009260 | 859 S | QRDPSEMRS | 0.997 | unsp | ETH_00009260 | 976 S | ALQLSPEAA | 0.991 | unsp | ETH_00009260 | 1098 S | ASGTSGFDS | 0.996 | unsp | ETH_00009260 | 1194 S | SVGLSAKEV | 0.991 | unsp | ETH_00009260 | 1617 S | PESASFKEG | 0.998 | unsp | ETH_00009260 | 1629 S | ASRSSAGDS | 0.998 | unsp | ETH_00009260 | 2062 S | TPPSSPLES | 0.997 | unsp | ETH_00009260 | 2095 S | CDRGSFKEV | 0.992 | unsp | ETH_00009260 | 2293 T | ATAATPKEA | 0.991 | unsp | ETH_00009260 | 2394 S | QTADSVDDK | 0.997 | unsp | ETH_00009260 | 2424 S | TEAASSRRE | 0.994 | unsp | ETH_00009260 | 2425 S | EAASSRREL | 0.997 | unsp | ETH_00009260 | 2437 S | VSRLSAAEK | 0.99 | unsp | ETH_00009260 | 602 S | FRPTSGRVH | 0.991 | unsp | ETH_00009260 | 704 S | QLLPSPKAD | 0.997 | unsp |