-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
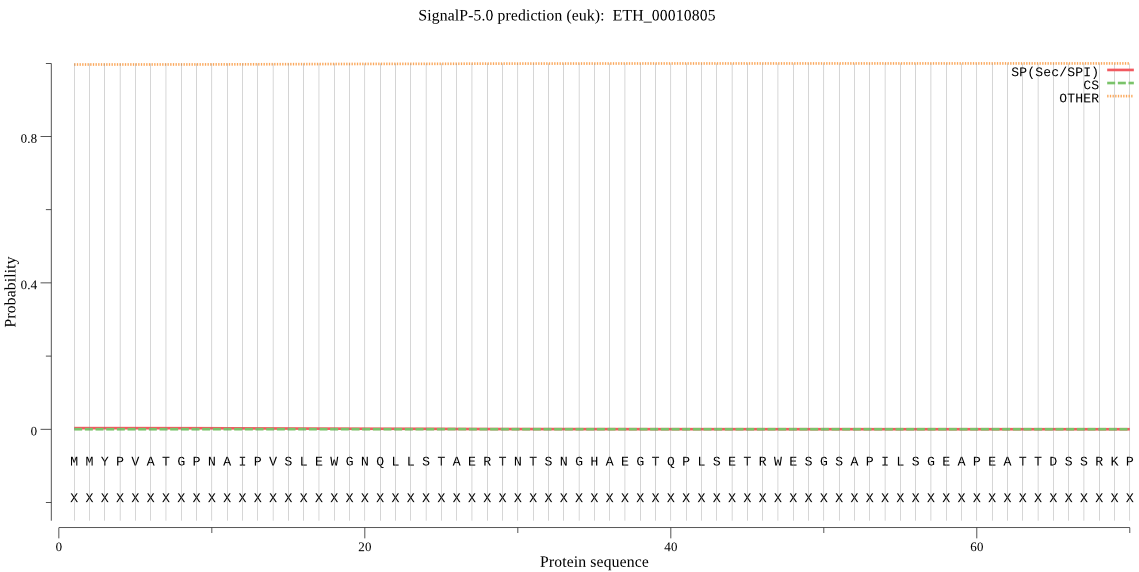
Fasta :-
>ETH_00010805
MMYPVATGPNAIPVSLEWGNQLLSTAERTNTSNGHAEGTQPLSETRWESGSAPILSGEAP
EATTDSSRKPAEATEDGCRSDATEIVPHWWRETCTKESFDQGSIVLCGCDGGASR
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/ETH_00010805.fa
Sequence name : ETH_00010805
Sequence length : 115
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.310
CoefTot : -0.784
ChDiff : -9
ZoneTo : 16
KR : 0
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 0.565 0.535 0.028 0.396
MesoH : -1.127 -0.630 -0.582 0.002
MuHd_075 : 8.644 13.242 2.791 1.150
MuHd_095 : 12.087 10.161 5.036 2.283
MuHd_100 : 19.773 15.085 6.041 3.573
MuHd_105 : 22.720 16.218 6.075 4.270
Hmax_075 : 7.787 10.063 -0.109 3.932
Hmax_095 : 13.300 11.600 2.041 4.960
Hmax_100 : 16.100 14.300 2.296 5.480
Hmax_105 : 15.517 17.033 2.645 5.810
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9828 0.0172
DFMC : 0.9814 0.0186
-
Fasta :-
>ETH_00010805
ATGATGTATCCGGTAGCGACAGGCCCTAATGCCATTCCAGTCTCCCTTGAGTGGGGAAAC
CAGCTCCTCAGCACTGCGGAAAGAACCAACACGAGCAATGGGCACGCGGAAGGGACTCAG
CCGCTTTCGGAAACGCGGTGGGAGTCTGGGTCGGCGCCGATCCTGAGTGGAGAGGCTCCG
GAAGCGACGACGGATAGCAGCAGGAAGCCTGCAGAGGCAACAGAAGACGGCTGTAGGTCA
GATGCGACAGAGATTGTGCCACACTGGTGGAGGGAAACTTGCACGAAAGAGTCTTTCGAC
CAAGGGAGCATTGTGTTATGTGGATGCGACGGTGGTGCTTCGCGTTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method |
|
ETH_00010805 | 67 S | TTDSSRKPA | 0.994 | unsp |


